EMI4_117_sm_Tables_S1-S4
advertisement

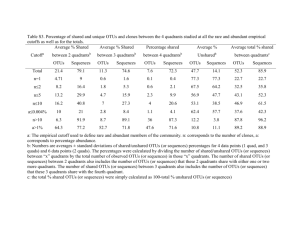
Supplementary data Table S1: Data related to the number of 16S rRNA PCR sequences. Bulk soil Rhizosphere C1BS C2BS C3BS C1R C2R C3R N° of 16S rDNA sequences 50609 56025 62022 42925 37358 52948 Maximum read length (bp) 362 323 315 322 308 304 Shortest read length (bp) 250 250 250 250 250 250 Average read length (bp) 265 266 266 264 265 264 Table S2: Estimate of the number of sequences required to reach the plateau OTUsMin* observed OTUs observed OTUs estimated (Chao1) Estimated number of sequences ** Low 95% Up 95% C1BS 8,281 9,872 18,365 484,079 942,174 C1R 4,886 5,264 9,738 264,745 458,860 C2BS 7,752 9,740 17,882 414,744 692,231 C2R 7,550 7,551 14,191 220,909 333,102 C3BS 5,178 6,830 12,570 399,458 670,394 C3R 5,619 6,843 12,995 444,985 923,719 * OTUs Min: OTUs observed when the numbers of sequences were normalized with the sample harboring the smallest number of sequences (C2R= 37358 sequences). ** the theoretical number of sequences required to reach the plateau was generated by combining the observed number of OTUs, the observed number of sequences and the theoretical number of OTUs (obtained with nonparametric Chao1 estimate ; lower and upper 95% confidence intervals). The theoretical number of sequences (bold) was obtained using linear equations as presented in figure S2 for the C2BS sample. Table S3: Number of sequences classified to be within known mineral weathering bacterial genera For each group, the number of sequences was evaluated at a 80% confidence threshold (bold upper value). The three independent samples are presented as follow: C1, C2 and C3. BS: bulk soil; R: rhizosphere. For each functional bacterial genus, a one-factor ANOVA (relative distribution values) and a Bonferroni-Dun test (p=0.05) were performed on the relative distribution values. Statistical analysis showed that there was significantly more Burkholderia (p=0.008) and Collimonas (p=0.045) in the rhizosphere than in the bulk soil. C1 C2 C3 BS R BS R BS R Burkholderia 431 1045 567 971 622 870 Collimonas 4 22 4 6 3 12 Mineral weathering bacteria Table S4: The ten most abundant genera in each of the six soils sampled as percent of total sequences from each soil. Bulk soil Rhizosphere Phylum Statistic Genus C1 C2 C3 C1 C2 C3 Acidobacterium 28.83 27.87 24.55 25.57 28.67 21.95 Acidobacteria n.s. Rhodoplanes 6.88 7.58 7.05 5.57 5.02 6.08 Alphaproteobacteria P=0.0132 (BS>R) Bradyrhizobium 3.34 3.54 4.22 4.06 3.42 4.56 Alphaproteobacteria n.s. Burkholderia 1.12 1.2 1.19 2.84 3.08 1.96 Betaproteobacteria P=0.008 (R>BS) Alterococcus 0.26 0.28 1.4 2.57 0.55 2.18 Gammaproteobacteria n.s. Chitinophaga 0.73 0.88 1.36 1.51 1.36 1.57 Bacteroidetes n.s. Mycobacterium 1.82 3.19 1.21 1.5 2.1 1.26 Actinobacteria n.s. Conexibacter 1.46 1.19 1.62 1.3 1.1 1.5 Actinobacteria n.s. Gemmatimonas 1.24 0.76 1.73 1.22 0.9 0.98 Gemmatimonadetes n.s. Spirillospora 0.55 0.41 0.77 0.92 0.76 0.91 Actinobacteria n.s. 0.17 0.2 0.17 0.19 0.23 0.95 Planctomycetes 0.87 0.79 1 0.8 0.55 0.69 Actinobacteria Planctomyces Acidimicrobium n.s. n.s. Titles and legends to fgures of the supplementary data: Figure S1 : Number of OTUs and Richness estimators a. Number of OTUs obtained. b. Shannon estimate; c. Chao1 estimate. The number of OTUs was determined using the RDP pyrosequencing pipeline. The non-parametric estimates were calculated using the EstimateS package. No significant differences were observed for any of these observed or theoretical indexes according to a one-factor ANOVA and the Bonferroni-Dun test (p=0.05). The solid circles and the open squares present bulk soil and Rhizosphere samples respectively. Figure S2: Presentation of the inverse values of the number of observed sequences and observed OTUs from C2BS sample. The last hundred values observed (OTUs and sequences) and presented in figure 1, were used to draw this graph. From this graph a linear regression was generated and a linear equation (y=ax+b) was obtained. The same approach was performed for each sample. For each sample, a linear equation was generated. Figure S3: Relative clone frequencies in Proteobacteria groups of the sequences from the oak rhizosphere and the surrounding soil. Soil samples are presented as follow at field-scale (C1, C2 and C3) and at micro-scale (BS: bulk soil and R: rhizosphere). One factor ANOVA (p=0.05) and a Bonferroni-Dun test performed on the relative distribution values evidenced that there was significantly more -,- and unclassifiedproteobacteria in the rhizosphere than in the bulk soil. -proteobacteria, p=0.0071 (R>BS); -proteobacteria, p=0.0482 (R>BS); unclassified-proteobacteria, p=0.0166 (R>BS).