Sulphur biochemistry - Plant Research International
advertisement

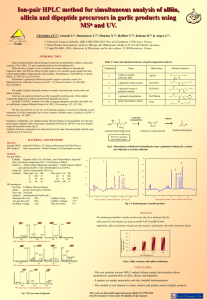
Workpackage number: WP4; Sulphur biochemistry Phase: 24-36 months Start date: Month 0 (February 1 2000) Completion date: Month 48 (January 31 2004) Current status: Ongoing Partners responsible: P2, P3 Person months per partner and total: P2: 66, P3: 132; total: 198 Already devoted person months per partner and total: P2:11.6, P3: 99; total: 110.6 Objectives The overall aim of this task is to understand the rate-limiting processes in the accumulation of alliin and its subsequent conversion to allicin. These include the biochemical steps in alliin synthesis and alliicin formation as well as sulphur transport within the plant. Three distinct objectives will be addressed using different but complementary approaches. The first objective is to identify the intermediates on the pathway(s) leading to synthesis of the major flavour precursor in garlic, alliin. This will involve detailed studies the biochemical pathways of sulphur compounds operating in garlic tissues, using specially synthesised labeled precursors. The second objective is to identify the developmental control points of cysteine sulphoxide synthesis and translocation. We will accomplish this by profiling and tracking of sulphur components in leaf and clove tissues throughout their growth and development. The third objective is to identify genes with altered expression in tissues with differences in levels of flavour compound pathway flux and genes involved in the conversion of alliin to alliicin. A number of approaches will be used for this, including differential display, cDNA library screening and related techniques. Any genes isolated from this part of the project will be made available for the genetic transformation programme in Task 2. Methodology and study materials Intermediates on the path(s) of synthesis will be identified by pulse-labeling techniques. Two potential pathways exist. For one the entry is through serine and will be studied using direct labeling of leaves and other organs with 14C serine. For the other, entry may be through glutathione and labeling will be by feeding roots with 35SO4 followed by analysis of the pattern of labeling in the leaves. Once the pathway has been outlined the next stage will be to examine subcellular localisation. The main sites of CSO synthesis within the whole plant and the stages of development where it occurs will be established by quantitative profiling of CSO and related compounds by HPLC, analysing leaf and bulb tissues during garlic growth in the field. Budgets will be constructed for individual and total sulphur compounds, along with other assimilates and quantitative information on the rates of synthesis and translocation of these compounds during the bulb cycle will be estimated. These studies will be complemented by pulse-labeling studies carried out in the glasshouse, in which whole plants will be labeled with 33SO4 at different stages during their development. Kinetic measurements on sulphur uptake and movement will indicate how transport patterns vary with development. Genes encoding enzymes in the biosynthetic pathway of alliin will be isolated by differential display. Potential genes will be located based by correlation of their expression in a range of tissues and conditions that show large differences in alliin synthesis. Candidate sequences will be used to screen a cDNA library to obtain full-length genes. Gene function will be assessed by comparison with database sequences for known enzymes and be confirmed by assaying the enzymic properties of the encoded proteins when expressed in E.coli and/or yeast. Clones encoding alliinase will be isolated from a garlic cDNA library and sequenced to identify different members of the gene family. The expression of genes encoding alliin synthesising enzymes and alliinases will be determined in different tissues at different stages of the garlic bulb cycle. Any novel alliinase or other gene sequences will be made available to the transformation programme for over-expression or targeted antisense. Deliverables Deliverable Status DP. 8: Analytical methods for labeling and analysis (P2, P3) DP. 9: A cDNA library from garlic (P2) DP. 16: Pathway intermediates identified (P3) DP. 17: First sulphur budget for garlic (P2) DP. 18: Clones for alliinase (P2) DP. 23: Publication on alliin biosynthesis and sulphur partitioning (P2, P3) DP. 24: Genes for key CSO synthesis enzymes (P2,P3) DP. 29: Publication on the characterisation of key enzymes in alliin biosynthesis (P2, P3, P5) DP. 35: Publication on the regulation of alliinase expression (P2, P3) DP. 36: Publication on the regulation of sulphur biochemistry in garlic (P2, P3, P5) = Achieved = delayed = future deliverable Milestones Year 1 Milestones Measurement of alkyl cysteine sulphoxides, pathway intermediates and gammaglutamyl peptides in bulbing and sprouting plants completed. Optimal conditions for pulse labeling of alliin established(P3). Experimental material planted and harvests begun (P2). Differential display of tissues exposed to different cultural conditions complete. (P3). cDNA library from actively synthesising tissues constructed (P2). Year 2 Milestones Radiolabelled intermediates identified using HPLC and HPLC-MS in comparison with chemically synthesised compounds (P3). First year harvests and analysis completed (P2). Second year experimental material planted. (P2). Differential display sequences and isolated and reanalysed by northern blots and in situ hybridisation, where possible, with other Allium sp. (P3) cDNA clones encoding alliinase isolated and sequenced (P2). Year 3 Milestones Analysis of pattern of labeling in later stage intermediates completed (P3). Analysis of second-year field experiment completed (P2). Whole plant labeling studies completed (P2). Publication on the developmental control of alliin synthesis submitted (P2). Purification of PCR amplified cDNA and sequencing completed (P3). Status Expression studies on alliinase clones initiated (P2). Year 4 Milestones Hypotheses on limiting steps in alliin synthesis tested. (P2, P3). PCR products, and full-length cDNA clones isolated (P2, P3). Heterologous expression in E. coli and/or yeast completed. (P3). = Achieved = delayed = future milestone Progress during the first reporting period. Good progress has been made during the first reporting period. All of the milestones were met in full and the two deliverables scheduled for the first year were delivered in full and on time. Progress against the relevant sections of the "Methodology and Study Materials" section is summarised below. Full details are given in the reports from Participants P2 and P3. "Intermediates on the path(s) of synthesis will be identified by pulse-labeling techniques. Two potential pathways exist. For one the entry is through serine and will be studied using direct labeling of leaves and other organs with 14C serine. For the other, entry may be through glutathione and labeling will be by feeding roots with 35SO4 followed by analysis of the pattern of labeling in the leaves." The flavour precursors alliin, isoalliin and methiin have been synthesised and also several possible intermediates in the path of synthesis of these compound. The identity of these compounds has been established by mass spectroscopy (MS) and nuclear magnetic resonance spectroscopy (NMR). Further compounds have been purchased, or obtained as gifts from members of the EU Garlic and Health consortium. The HPLC behaviour of these compounds has been established in our analytical system. The method of extraction from garlic tissues, and separation by HPLC of the flavour precursors and possible intermediates has been simplified and the repeatability of the methods confirmed. The formation of alliin by garlic tissue cultures from the intermediates, allyl cysteine and allyl thiol, indicated that callus was a suitable tissue to study the path of synthesis of alliin, the major flavour precursor. Synthesis of isoalliin by callus after incubation with carboxypropyl cysteine showed that the tissue was also capable of synthesising isoalliin and could therefore be used to study the synthesis of this minor flavour precursor in garlic. "The main sites of CSO synthesis within the whole plant and the stages of development where it occurs will be established by quantitative profiling of CSO and related compounds by HPLC, analysing leaf and bulb tissues during garlic growth in the field." The need to profile sulphur accumulation and re-mobilisation in different tissues of garlic required the development of a cultivation method that allows sulphur inputs to be managed and labeled sulphur to be added or removed from the growing garlic. This has been achieved by growing the garlic varieties in a hydroponic system rather than in soil. During the reporting period a new hydroponic system has been developed and tested by growing garlic plants successfully to the bulbing stage. Test Cysteine sulphoxide determinations have been carried out on cloves from a single bulb grown hydroponically using the HPLC methods developed by partner P2. Alliin is the major CSO at nearly 80% of the total CSOs in garlic clove tissues. "Genes encoding enzymes in the biosynthetic pathway of alliin will be isolated by differential display. Potential genes will be located based by correlation of their expression in a range of tissues and conditions that show large differences in alliin synthesis." We have established a differential display method that displays a highly reproducible banding pattern from independent RNA extractions. Several experimental difficulties were identified and overcome successfully. We have used both cut leaf sections and garlic tissue culture as sources of mRNA. In order to establish conditions under which tissues will differentially synthesise alliin it has been necessary to establish cultures of garlic growing in vitro. This has been achieved. Callus cultures of variety Printanor have been initiated and are being maintained regularly. In addition, attempts were made to initiate cultures from variety Messidrôme but progress has been slow. Recently the inhibition on root production has been overcome. "Clones encoding alliinase will be isolated from a garlic cDNA library and sequenced to identify different members of the gene family." In order to isolate alliinase gene sequences from garlic it is necessary to construct a cDNA library from tissues actively expressing the enzyme. Two cDNA libraries have been produced, one from bulb and one from leaf material. Both libraries have in excess of 2x106 plaque forming units and an estimated size range of 400 - 3000+ base pairs. These will be used to isolate alliinase sequences from garlic and are available for the isolation of full-length clones of genes isolated in differential display experiments. Progress during the second reporting period. Good progress continues to be made in the work package. All of the milestones and deliverables for the second year were achieved. Detailed accounts of the work in this workpackage are contained in the individual reports from partners P2 and P3. The following summarised progress against the Methodology and Study Materials. "Intermediates on the path(s) of synthesis will be identified by pulse-labeling techniques." Exposure of the garlic tissue to allyl thiol led to the accumulation of both allyl cysteine and alliin. Allyl cysteine, when taken up by the tissue culture, was readily oxidized to alliin. Although the amounts of propiin are generally very low in garlic, the callus was still capable of making the conversion from propyl thiol to propyl cysteine and propiin. Under low sulphur conditions only the final stage of the pathway is found after feeding the same intermediates i.e. alliin, isoalliin or propiin. Interestingly, onion callus, which does not normally accumulate alliin, formed alliin when exposed to either allyl thiol or allyl cysteine. Propenyl cysteine and propyl cysteine were oxidized to their respective isoalliin and propiin end products in both garlic and onion callus. Most of these conversions occurred after 5 days incubation. For the system studied the generalised reaction of: alk(en)yl thiol - alk(en)yl cysteine - alk(en)yl cysteine sulphoxide can be carried out by callus. This may indicate that a generalised alk(en)yl cysteine synthase and alk(en)yl cysteine oxidase enzyme(s) exist in garlic and onion tissues. These feeding experiments have given new insights into potential key steps in the synthesis of alliin, in particular indication the potential importance of cysteine synthase in the pathway. "The main sites of CSO synthesis within the whole plant and the stages of development where it occurs will be established by quantitative profiling of CSO and related compounds by HPLC, analysing leaf and bulb tissues during garlic growth in the field." To profile sulphur accumulation and re-mobilisation in different tissues of garlic we grew garlic plants hydroponically in a defined medium using pot-grown plants as a control. Plants were been harvested at regular intervals and the fresh weight, dry weight, total sulphur, total nitrogen, total carbon, total solubilised protein and the cysteine sulphoxide profile determined for clove leaf and root. Measurements were made on two genotypes, Printanor and Messidrome. This data has allowed us to build up for the first time, a picture of the dynamic uptake, allocation and redistribution of carbon, nitrogen and sulphur through the garlic life cycle. We can identify four stages of development from clove planting. The first a period of slow growth and establishment sustained by resources within the clove. The second is a period of rapid and active growth of leaves and root. It is during this phase that the bulk of sulphur and nitrogen is taken up by the plant and the bulk of CSOs are formed. The third phase is bulb formation, during which sulphur (including CSOs) and nitrogen are redistributed to the new cloves in the bulb. Additional carbon continues to be assimilated during this period and accumulates in the bulb. The last phase is maturation where the leaves senesce and die and bulb formation is complete. We will look in more detail at the redistribution of assimilates in the next year. "Genes encoding enzymes in the biosynthetic pathway of alliin will be isolated by differential display. Potential genes will be located based by correlation of their expression in a range of tissues and conditions that show large differences in alliin synthesis." Following on from the previous year, a differential display method that displayed a highly reproducible banding pattern from independent RNA extractions was developed. Twelve differentially expressed cDNAs were identified that showed no function in flavour precursor biosynthesis. We concluded that a different approach was essential to continue this work successfully. A major objective of the workpackage is to identify key enzyme(s) in the pathway(s) leading to alliin biosynthesis. Based on a comprehensive review of recent literature and the feeding experiments described above the hypothesis that cysteine synthase (CSase) and serine acetyltransferase (SATase) are key enzymes in flavour precursor biosynthesis was developed and work put in place to test this. Five potential CSase cDNA fragments were successfully PCR amplified. Sequence analysis revealed three distinctly different nucleotide sequences that when aligned fell into two groups with significant homology with plant CSases. In addition, three identical sequences with strong homology to SATase were isolated. This work was complemented by a biochemical approach to purify a CSase enzyme fraction from Garlic. A 34 kDa protein was isolated that contained peptides consistent with a plant cysteine synthase. This enzyme may be the one responsible for the conversion of a substrate, possibly an allyl thiol, to allyl cysteine. Primers designed from these peptides should allow further characterisation of this enzyme. "Clones encoding alliinase will be isolated from a garlic cDNA library and sequenced to identify different members of the gene family." Alliinase sequences were amplified from both the bulb and root cDNA libraries described in last year's report, using the polymerase chain reaction. The primers were based on alliinase sequences from onion. The garlic alliinase fragments were cloned in to a plasmid vector and sequenced. At least five different alliinase sequences are present in the variety Messidrome. Sixteen of the twenty-one clones, from both bulb and leaf cDNA libraries were the same sequence. This suggests that this is the dominant alliinase mRNA in this variety. All leaf-derived sequences, with one exception, were the same. Bulb-derived sequences fell in to four groups. Sequences identical to the major sequence in the leaf were most prominent suggesting that this sequence may encode the dominant alliinase for the whole plant. The bulb cDNA library was probed using one of the cloned alliinase sequences. Fifty four plaques were isolated and will be sequenced to identify full length clones of the gene. Progress in the third reporting period Milestone: 2002: Analysis of pattern of labeling in later stage intermediates completed. Callus was exposed to an enlarged range of single substrates at 1 and 10mM concentration (serine, cysteine, glutathione, 2-carboxy propyl cysteine, 3-carboxy propyl cysteine, allyl alcohol, propyl alcohol, vinylacetic acid, methacrylic acid and cystathione) and reduced times of incubation. (1 and 4 days). Analysis of this experiment was halted when it became apparent that the callus was very variable such that supposed undifferentiated callus showed signs of differentiation, thus invalidating the significance of any results obtained. This variation in callus has been reduced by selecting one rapidly growing callus as a source for all subsequent subcultures. The aim was to repeat the precursor feeding with garlic callus grown from this single selected clone so as to establish whether the serine pathway was functional in garlic. Undifferentiated garlic callus was maintained on a phytogel medium and a sulphur depleted medium (all sulphate compounds replaced with chloride). A range of potential precursors, synthesized or purchased, was incorporated in callus medium prior to incubation. The production of any alk(en)yl cysteine conjugate and/or alk(en)yl cysteine sulphoxide was measured by HPLC after extraction of the callus. The precursors used were allyl thiol, propyl thiol, allyl cysteine and propyl cysteine all at a concentration of 10mM. The tissue was sampled at 2, 6 and 13 days. There was an increase in alliin, propiin, allyl cysteine and propyl cysteine when the relevant intermediates were used which indicated the network nature of the metabolic pathways. There were no obvious differences between the sampling times. It appeared that thiols provided an intermediate for the synthesis of alliin or propiin confirming the possibility of the existence of this pathway. Milestone 2002: Analysis of second-year field experiment completed. An experiment was carried out to examine the patterns of suphur uptake and partitioning as a repeat of a similar experiment carried out in 2001. In addition, labeled sulphur was fed to developing garlic plants to attempt to determine the patterns of partitioning of organic sulphur during garlic growth and clove formation. Samples from this experiment are still being analyzed and the full data set will be available in the early part of the fourth reporting period. From the data so far available (see report of partner P2), the pattern of sulphur accumulation was similar to that in the previous years, although some detailed aspects of the timing of changes in development were observed, mainly reflecting variability in the environmental conditions experienced year on year. Milestone 2002: Whole plant labeling studies completed. As described in the previous section, an experiment to determine the fate of labeled sulfur fed to garlic plants at different stages of development was carried out. The samples are undergoing analysis and the results will be available in the early pray of the next reporting period (see report of partner 2 for details). Milestone 2002: Publication on the developmental control of alliin synthesis submitted. DP. 23: Publication on alliin biosynthesis and sulphur partitioning. The paper is in preparation but completion requires the data from experiments on developmental control of alliin synthesis, which are still being analyzed. Milestone 2002: Purification of PCR amplified cDNA and sequencing completed. DP 24: Genes for key CSO synthesis enzymes This milestone concentrated on genes for Cysteine synthase (CSase) and serine acetyl transferases (SATases). Screening for a novel CSase that exhibits S-allyl-CSase activity led to the isolation of four clones encoding cysteine synthase. 1. GCS1 – a potential pseudogene 2. GCS2 – a potential choroplastic type 3. GCS3 – a cytosolic type 4. GCS4 – a potential S-allyl-CSase. Functional studies n these genes were attempted using an inducible transgenic expression system based on ethanol induction. For technical reasons, this strategy was unsuccessful and alternative inducible systems are being investigated. A plasmid cDNA library was constructed from combined total RNA extracted from garlic clove, leaf and root organs. An aliquot of the cDNA library was enriched with a PCR product derived from a cDNA fragment with the aim to detect at least one SATase isoform. Among the 1.6 x 10 4 colonies screened one full-length SATase cDNA was identified. Functional studies were attempted as described above, but for the same technical reasons, these were unsuccessful. Milestone 2002: Expression studies on alliinase clones initiated. DP 35; Publication on the regulation of alliinase expression. Alliinase expression was measured using a RT-PCR approach, using the 18S rRNA as an internal standard. Alliinase expression in both clove and leaf tissue did not vary significantly with stage of development. This suggests that the allinase genes may be constitutively expressed in garlic. This is in contrast to onion, where some developmental regulation has been observed. Garlic alliinase may have defensive role as it's primary function, supported by the observation that it is only located in the vacuoles of bundle sheath cells and not in the abundant storage mesophyll cells. This is in contrast to the distribution of alliinase in onion, which is found in all cell types. Aliinase is extremely abundant in garlic tissue consisting of at least 10% of the total clove protein and it is therefore possible that it also functions as a storage protein. These experiments were initiated in 2002 and are still in progress. The paper will be written and submitted when experiments are completed in 2002. DP 36: Publication on the regulation of sulphur biochemistry in garlic. This paper will pull together work on the biochemistry on alliin synthesis undertaken by P3 and the developmental and sulphur budget studies by P2. These will be completed in early 2003 and the paper will be prepared at that time.