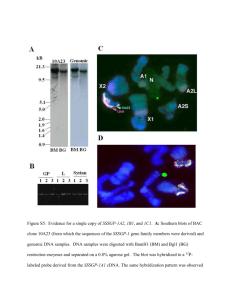
Experimental Procedures for Grant Write-Up (Illumina
advertisement

Experimental Procedures Illumina BeadChip analysis of genotyping will utilize the 660 quad BeadChip. Each SNP is represented on these chips by on average 30 beads. The studies will be carried out by submitting genomic DNA samples to the Keck Microarray Resource (http://keck.med.yale.edu/microarrays) which is a full service, cost recovery unit that carries out sample processing, hybridization, and data analyses. The quality of genomic DNA will be evaluated by A260/A280 ratio, which should be at least 1.9, and by electrophoresis on either the Agilent Bioanalyzer or Invitrogen E-gels. For each experimental condition 1 ug of very high quality genomic DNA will be provided to the facility which will then carry out the whole genome amplification, hybridization, and data analysis as described below. Preparation of amplified DNA for hybridization onto Illumina BeadChips will follow the recommended Illumina protocol. Whole genome amplified DNA will be prepared utilizing the Infinium II assay (Illumina). Approximately 1 mg of the amplified sample will be enzymatically fragmented, precipitated, and resuspended in a proprietary buffer RA1. Samples will be loaded onto BeadChips using automation in preparation for a 16 h hybridization at 48C. An automated single-base extension reaction and signal amplification will take place following hybridization using Infinium II kit reagents. The BeadChips will be further prepped by coating with a UV protectant and scanned using the IScan. Scanned output files will be analyzed and call rates calculated using Illumina BeadStudio. The quality of the data will be evaluated by both sample dependent and sample independent controls. Specifically, efficiency of target removal, non-specific binding, and appearance of cross-contamination will be examined.