iTraq_labeling_PvB
advertisement

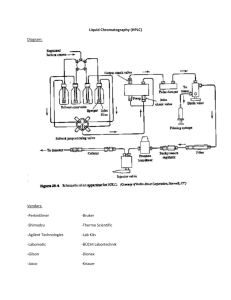
iTraq labeling - Put freeze-dried samples on ice (use right eppendorf tubes with safe-lock and plastic that does not come to solution at low pH) - Dilute 1.0 M trietylamoniumbicarbonate buffer 1:1 with water 0.5 M dissolution buffer, put on ice - Add 126 ul dissolution buffer per vial RapiGest, vortex and put on ice - Add 29 ul RapiGest (to denature proteins) to each sample, until protein pallet is completely dissolved, spin short and store at RT for 10 min - Add 2 ul reducing buffer to each sample, vortex, spin short, and put in shaker incubator at 55 C, 400 rpm for 1h - Add 1 ul blocking buffer, vortex, spin short and store at RT for 10 min - Add 55 ul dissolution buffer to each vial of trypsin - Add 10 ul trypsin to each sample, vortex, spin short, and incubate o/n (12 – 16h) in shaker incubator at 37 C, 400 rpm - Next day, spin samples down short and to each sample ad 80 ul of HPLC purified isopropanol - Then add a volume of iTraq label corresponding to one unit (U) according to ABI. This is ~18ul. Vortex, spin down and leave at RT on round vortexer for 2h. - Spin down short and pool all samples from the same series - Dilute 25% TFA 5x to 5% and add 25 ul 5% TFA per sample to the pooled samples (so 200 ul for 8 samples). This is to break down the RapiGest - See if pH is 3.0 using pH paper and vortex at RT for 30 min - Speed-vac the pooled samples o/n - Next day, put dried pallet in -80 C. -----For common reference (CR) mix: Reconstitute dried pallet in 600 ul of 0.5 M dissolution buffer, vortex to completely dissolve, and divide over 6 eppendorf tubes (100 ul per tube). Speed-vac (2.5h) and store at -80 C (extra speed-vac step is to get rid of TFA completely to store CR samples over a longer time) -----To start HPLC and iTraq: (Before loading samples, calibrate, test (with BSA digest) and wash HPLC) - Add 170 ul Buffer (10mM KH2PO4, 20% CAN) to CR pallet and reconstitute by vortexing. Add this to sample mix and reconstitute by vortexing - Alternatively, when protein amount is very high, load two times on HPLC (the reconstitute sample en 2 x 170 ul). Now you have two samples per fraction. This will increase the separation of the samples - Start Chromeon software - In window HPLC runs: open file with runs Pieter - Add a run by clicking on last run and pres down - Add name of run (ie Pieter Set1), sample volume (160 ul) and position number where you place your sample in the rotor - Check program by double clicking on it and check if fractions are collected in the correct tubes at the correct time point by the fraction collector - Put 1,5 ml tubes at correct places in fraction collector (collects 200 ul per minute) - Ad 170 ul sample to HPLC tube and put in rotor at position number - Save file - Click on edit batch button, click file and load file ‘Pieter’, perform ready check and if ok click start - Monitor run in HPLC window - After run, collect and label fractions (1-40), speed-vac and store in -80 for fractionation C18 C18 – colum: - Wash plate with Et-OH, water, soap, water, Et-OH and polish with metal polish - Put plate in spotter with flat corner to upper left side - Reconstitute HPLC1 fraction in 40 ul 0.1% TFA and put 20 ul in HPLC tube. Start with fraction in middle of the second peak and work your way out to where there is hardly any peptide. Refreeze rest of the sample - Alternatively, when first HPLC was done in two steps, reconstitute one sample from each fraction in 20 ul and load all 20 ul in HPLC tube (you have the second tube from the fraction as a backup). - Setup 2nd HPLC program; load 10 ul, use program Roel 120, name sample and add number of plate fractions are spotted on - When adding multiple plates at a time to run overnight, then also add a blank run and a stop run - Make sure spotting program is setup correctly - Click run and perform performance check and start - After spotting store plate in labeled box in drawer - On these plates add 0.4 ul of calibration peptides in matrix on calibration spots, and let dry (~5 min) Load plate in MS/MS MS/MS - File new spot set create new name this with date - Use defined spot templates select ‘Roel LC 1-384, (when two fractions are spotted on one plate select Roel LC 1-2) and click select - Job template Map Roel methods 384 2KV apply - Save - Plate load plate MS/MS analysis MS/MS spectra are uploaded by GPS software to the SWISS Prot and NCBI database. Swiss prot hits serve as standards and additional hits in NCBI are added to this. - Start GPS: login: admin, password: admin - New project (Pieter) Right click Pieter: New sample set (Name: Set 1 Fr18) - Workflow: LC MALDI - Select spot sets AK065 (MS pc) Pieter fractie - Save - Add new analysis to sample set; right click sample set and add, name according to database (ie Swiss prot, rat) - Type: MS/MS - Template: Swiss prot, 8 plex, Rat (you can check if the standard settings of the analysis is correct) - Select Job runs select MS/MS runs (2nd MS step) corresponding to correct fraction - Save - Submit Job - Add new analysis to sample set where you do the same for the NCBI database (you can run maximum two analysis parallel) You can check the progress in the job manager - After run is complete open result browser and check protein summary - Click the MS/MS summary and sort by observed mass and copy in excl sheet - Do the same for the NCBI data and paste these in the second tab of the same exell sheet. Name this sheet according to set and fraction number and save in GPS folder. - To recover iTraq data corresponding to the peptides, open program TS2Mascot, open fraction data. Select 2. job run MS/MS data and save peak list in mgf data folder - open ‘itraq analysis automatically’ folder and past mgf file in this folder (do not drag). And click batch.pl application - when done, open the mgf.itraq file in excel and save as excel sheet in iTraq folder.Sort by precursor mass and paste in GPS file in Swiss and NCBI sheet. Check if precursor masses from GPS file and MGF file correspond correctly and delete colums of spot labels and precursor so that the iTraq data are directly after the accuisition time collum. Save the file.