Proteomic characterization of a conditional lethal in
advertisement

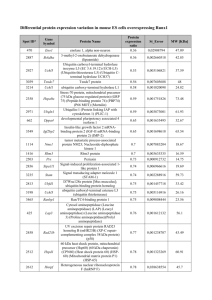
Supplementary material to Pedersen et al.: Proteomic characterization of a temperature sensitive conditional lethal in Drosophila melanogaster. Titles and legends to figures in supplementary material Figure S1 Unused ProtScore for all identified proteins in the 5 iTRAQ experiments. An Unused ProtScore of 1.3 was used as a threshold in all experiments. The Unused ProtScore estimates the confidence by which a protein can be validated, using only peptide evidence that is not better explained by a higher ranking protein. An Unused ProtScore of 1.3 corresponds to an identification confidence of 95%, and an Unused ProtScore of 2.0 corresponds to a confidence of 99%. The Unused ProtScore and the identification confidence are related according to following equation Unused ProtScore = - log (1 percent confidence/100). Figure S2 Protein sequence coverage (%) for all identified proteins in the 5 iTRAQ experiments. Figure S3 Expression levels of a subset of proteins identified in experiment 3. Proteins related to mitochondria are shown in A) and some of the other differentially regulated proteins in B). Expression level for each protein at the different temperatures is relative to the IC-line at the permissive temperature. Stars indicate significance level (* = P < 0.05, ** = P < 0.01, *** = P < 0.001) relative to the IC-line at the permissive temperature (20°C). Level of IC-line at the permissive temperature is indicated by the dashed horizontal line. 2 Figure S1 100 Unused ProtScore 80 60 40 Mean = 9.2 Median = 4.4 20 0 All identified proteins 3 Figure S2 100 % protein coverage 80 60 Mean = 39.36% Median = 36.27% 40 20 0 All identified proteins 4 0.0 5 * * *** B Adh related Turandot A * 1.8 IC-line at 29°C L-line at 20°C L-line at 29°C Turandot C ** A Cytochrome c oxidase mitochondrial precursor ** * * 2.0 Putative ATP synthase f chain, mitochondrial ATP synthase subunit d CG3731 ATP synthase alpha chain mitochondrial precursor *** *** *** *** *** * *** 1.4 Stress sensitive B ** *** 1.2 Porin Relative protein regulation (relative to IC20) Figure S3 1.6 1.0 0.8 0.6 Table S1 Gene-Enrichment-Analysis of the significantly differentially expressed proteins between the L-line and the IC-line at permissive temperature (20°C) from experiment 3. The number of genes identified within specific groups, the percentage of total genes within groups and the EASE score P-value for the Gene Ontology Themes ‘Biological Process’, 6 EASE score P-value Biological Process No significant biological processes Molecular Function Transporter activity Transmembrane transporter activity Active transmembrane transporter activity Cellular Compartment Mitochondrial part Mitochondrion Mitochondrial membrane Mitochondrial envelope Lipid particle Organelle envelope Envelope % of group # of genes ‘Molecular Function’ and ‘Cellular Compartment’. 9 7 5 22.0 17.1 12.2 1.20E-02 2.70E-02 3.70E-02 11 12 6 6 8 6 6 26.8 29.3 14.6 14.6 19.5 14.6 14.6 8.80E-04 4.40E-03 2.70E-02 3.50E-02 3.70E-02 3.90E-02 3.90E-02 Table S2 Gene-Enrichment-Analysis of the significantly differentially expressed proteins between the L-line and the IC-line at restrictive temperature (29°C) from experiment 3. The number of genes identified within specific groups, the percentage of total genes within groups and the EASE score P-value for the Gene Ontology Themes ‘Biological Process’, 7 EASE score P-value % of group Biological Process Cellular lipid metabolic process Lipid metabolic process Monocarboxylic acid metabolic process Fatty acid metabolic process Coenzyme metabolic process Fatty acid beta-oxidation Fatty acid oxidation Cofactor metabolic process Organic acid metabolic process Carboxylic acid metabolic process Cellular carbohydrate metabolic process Molecular Function Oxireductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor Oxireductase activity Oxireductase activity, acting on the CH-OH group of donors Transferase activity Cellular Compartment Intracellular membrane-bound organelle Membrane-bound organelle Mitochondrion # of genes ‘Molecular Function’ and ‘Cellular Compartment’. 10 10 9 6 11 4 4 11 12 12 11 18.5 18.5 16.7 11.1 20.4 7.4 7.4 20.4 22.2 22.2 20.4 9.00E-05 7.10E-04 2.10E-03 1.30E-02 1.30E-02 1.40E-02 1.40E-02 1.80E-02 2.00E-02 2.00E-02 4.60E-02 8 17 14.8 31.5 1.10E-02 2.70E-02 8 12 14.8 22.2 4.10E-02 4.20E-02 18 18 13 33.3 33.3 24.1 4.10E-03 4.10E-03 1.60E-02 Table S3 List of differentially regulated proteins in response to the restrictive temperature in the lethal-line (L-line). P-value from t-test and FlyBase accession number are listed. ‘Quantification’ column indicates # of peptides used in the t-test, ‘Identification’ column indicates the # of unique but potentially overlapping peptides identifying the protein. Proteins involved in oxidative phosphorylation (ox) and associated to the mitochondria (m) are indicated m ox m m ox m m ox m m m 560 2439 1556 104 93 176 49 362 284 1249 76 278 37 629 539 11 593 237 124 60 87 43 24 22 15 18 25 28 74 17 30 3 44 44 2 70 18 13 Cytological position 17A9-17A9 35B3-35B3 80A-80B 88E12-88E13 88E13-88E13 91F1-91F1 36B1-36B1 98F12-98F13 9E10-9F1 102D1-102D1 1B4-1B4 85B7-85B7 14B8-14B8 66C8-66C8 85A5-85A5 6C5-6C5 46B4-46B4 32B1-32B1 44B3-44B3 L29/L20 (115/117) ox P value 2.15E-28 1.08E-22 8.78E-10 1.91E-09 2.18E-08 3.16E-08 6.61E-08 2.14E-07 3.83E-07 6.97E-07 9.50E-07 5.91E-06 6.57E-06 1.95E-05 2.67E-05 4.40E-05 8.15E-05 1.35E-04 2.56E-04 L29/IC20 (115/116) M L20/IC20 (117/116) Ox IC29/IC20 (114/116) Transferrin 1 Adh-related Sarcoplasmic calcium-binding protein 1 Tropomyosin 1 Tropomyosin 2 ATP synthase, subunit d Myosin heavy chain Phosphoglyceromutase Stress-sensitive B ATP synthase-β Cytochrome P450 4g1 CG11963 CG3560 Isocitrate dehydrogenase CG8036 CG3192 CG1516 Porin Obp44a FlyBase ID FBgn0022355 FBgn0000056 FBgn0020908 FBgn0003721 FBgn0004117 FBgn0016120 FBgn0086783 FBgn0014869 FBgn0003360 FBgn0010217 FBgn0010019 FBgn0037643 FBgn0030733 FBgn0001248 FBgn0037607 FBgn0029888 FBgn0027580 FBgn0004363 FBgn0033268 Identification Protein name Quantification based on the DAVID Gene-Enrichment-Analysis. 1.131 1.040 1.053 0.769 0.748 1.209 1.026 0.934 1.017 1.037 1.127 0.998 1.201 0.974 1.088 1.262 0.921 1.061 0.948 0.826 1.079 0.926 0.895 0.844 1.390 2.030 0.921 1.410 1.142 1.602 0.832 1.655 0.939 0.931 2.232 0.815 1.424 1.141 1.277 1.286 1.044 0.964 0.967 1.345 1.244 1.007 1.303 1.087 1.188 0.951 1.561 0.846 0.944 2.054 0.837 1.365 0.963 1.548 1.200 1.131 1.073 1.136 0.979 0.622 1.098 0.924 0.961 0.736 1.142 0.957 0.901 1.015 0.934 1.031 0.966 0.841 Error factor 1.042 1.025 1.020 1.095 1.085 1.111 1.139 1.043 1.051 1.027 1.142 1.045 1.107 1.032 1.036 1.198 1.038 1.056 1.062 Myosin light chain alkali Larval visceral protein H CG9090 CG4769 Stress-inducible humoral factor Turandot A Esterase 6 precursor CG9029 CG9468 CG3321 CG18815 Senescence marker protein-30 CG3731 Accessory gland-specific peptide 26Aa Probable phosphoserine aminotransferase CG10512 Troponin C 41F Ribosomal protein S18 Histone H4 replacement Malic enzyme Cytochrome c oxidase polypeptide Va Ribosomal protein L12 CG16712 CG33138 Stress-inducible humoral factor Turandot C ATP synthase B chain, mitochondrial precursor CG8343 FBgn0002772 FBgn0002570 FBgn0034497 FBgn0035600 FBgn0028396 FBgn0000592 FBgn0031746 FBgn0032069 FBgn0038224 FBgn0042138 FBgn0038257 FBgn0038271 FBgn0002855 FBgn0014427 FBgn0037057 FBgn0033027 FBgn0010411 FBgn0013981 FBgn0002719 FBgn0019624 FBgn0034968 FBgn0031561 FBgn0053138 FBgn0044812 FBgn0019644 FBgn0040502 ox m m ox m ox m ox m ox m 4.11E-04 4.70E-04 5.27E-04 5.31E-04 1.12E-03 1.20E-03 1.38E-03 1.68E-03 2.38E-03 2.73E-03 3.27E-03 3.28E-03 3.66E-03 3.71E-03 3.80E-03 4.79E-03 4.87E-03 5.00E-03 5.14E-03 5.21E-03 5.88E-03 6.70E-03 7.14E-03 7.44E-03 7.63E-03 7.66E-03 9 18 17 61 41 70 78 75 25 20 27 79 81 71 8 19 58 25 6 308 90 10 36 151 45 31 19 3 9 10 8 10 15 19 9 2 6 12 10 14 5 4 8 7 2 30 10 4 4 28 7 7 2 98A14-98A15 44D1-44D1 56F15-56F16 64C13-64C13 93A2-93A2 69A1-69A1 26A1-26A1 29F1-29F1 88B4-88B4 68D1-68D1 88D2-88D2 88D6-88D6 26A1-26A1 99A1-99A1 78C2-78C2 41F2-41F3 56F11-56F11 88C9-88C9 87C6-87C7 86F9-86F9 60B7-60B7 24B3-24B3 49F4-49F4 93A2-93A2 67C5-67C5 42A13-42A13 1.284 1.044 1.092 1.253 1.304 1.099 1.159 0.630 1.114 0.841 1.179 1.158 0.879 0.772 1.016 1.002 0.923 0.385 0.988 1.147 0.984 0.719 0.886 1.971 1.083 1.091 1.221 1.257 1.484 1.932 0.482 1.387 0.869 1.911 1.396 0.885 1.186 1.501 0.962 0.486 0.845 1.157 1.200 0.471 0.875 1.463 1.291 1.014 1.158 0.383 1.622 0.795 0.909 0.810 1.304 1.779 0.455 2.021 1.407 0.678 1.396 1.068 1.131 1.439 1.063 0.537 1.054 0.900 0.930 0.471 0.986 1.442 0.931 0.942 0.933 0.499 1.396 0.486 0.759 0.655 0.881 0.930 0.949 1.473 1.602 0.361 1.005 1.205 0.962 0.964 1.105 1.102 1.254 0.786 0.779 1.007 1.128 0.996 0.733 0.939 0.811 1.298 0.873 0.621 1.209 1.191 1.103 1.154 1.161 1.115 1.208 1.311 1.072 1.182 1.111 1.110 1.107 1.244 1.136 1.142 1.159 1.391 1.049 1.097 1.331 1.132 1.075 1.255 1.172 1.188 Table S4 Proteins significantly differentially expressed between the L-line and the IC-line at the permissive temperature (20°C) Protein name Larval serum protein 2 Glutamate oxaloacetate transaminase 2 MET75Cb protein precursor ATP synthase alpha chain, mitochondrial precursor Voltage-dependent anion-selective channel CG31883 CG31305 CG6287 Stress sensitive b Heat shock protein 60 Probable acyl-CoA dehydrogenase, medium-chain specific, mitochondrial precursor CG8626 CG18067 Neural conserved at 73EF Serine protease inhibitor Ejaculatory bulb specific protein III precursor CG15369 CG30415 CG8343 CG9914 Aldehyde dehydrogenase CG10924 Thiolester containing protein IV Succinyl-CoA ligase alpha-chain, mitochondrial precursor CG10664 CG11151 Cytochrome P450 4g1 FlyBase ID FBgn0002565 FBgn0001125 FBgn0028415 FBgn0011211 FBgn0004363 FBgn0032122 FBgn0037913 FBgn0032350 FBgn0003360 FBgn0015245 FBgn0035811 FBgn0034152 FBgn0034512 FBgn0010352 FBgn0028986 FBgn0011695 FBgn0030105 FBgn0250838 FBgn0040502 FBgn0030737 FBgn0012036 FBgn0034356 FBgn0041180 FBgn0004888 FBgn0032833 FBgn0030519 FBgn0010019 10 L20/IC20 (117/116) 0.73 0.83 1.14 1.09 1.16 0.49 1.13 0.85 1.17 0.87 0.86 0.81 0.83 0.91 1.17 1.12 1.22 1.37 0.90 1.14 0.94 0.83 1.27 0.92 1.12 1.12 1.14 P-value 5.00E-16 1.24E-05 1.81E-05 2.35E-04 3.49E-04 4.00E-04 4.22E-04 5.31E-04 7.45E-04 8.65E-04 9.34E-04 1.20E-03 2.01E-03 2.65E-03 3.20E-03 3.27E-03 3.30E-03 4.76E-03 6.24E-03 6.45E-03 8.52E-03 1.01E-02 1.08E-02 1.34E-02 1.40E-02 1.66E-02 1.87E-02 Error factor 1.06 1.09 1.06 1.04 1.08 1.45 1.07 1.08 1.09 1.08 1.09 1.12 1.06 1.06 1.10 1.07 1.09 1.20 1.06 1.09 1.05 1.15 1.19 1.07 1.06 1.10 1.11 CG6188 ATP synthase , subunit d CG11255 Sorbitol dehydrogenase Dihydropteridine reductase Adh-related Fructose-bisphosphate aldolase CG7966 CG12321 Phosphatidylinositol transfer protein CG12082 CG15616 40S ribosomal protein S20 Guanine nucleotide-binding protein gamma-e subunit FBgn0038074 FBgn0016120 FBgn0036337 FBgn0022359 FBgn0035964 FBgn0000056 FBgn0000064 FBgn0038115 FBgn0038577 FBgn0026158 FBgn0035402 FBgn0034153 FBgn0019936 FBgn0028433 1.30 1.18 0.80 0.84 1.18 1.39 1.03 1.34 1.13 0.81 0.98 0.86 0.82 1.45 1.87E-02 1.97E-02 2.01E-02 2.07E-02 2.22E-02 2.33E-02 2.69E-02 2.82E-02 3.00E-02 3.29E-02 3.37E-02 3.64E-02 3.75E-02 4.90E-02 1.10 1.15 1.19 1.14 1.15 1.31 1.03 1.27 1.07 1.15 1.01 1.15 1.19 1.44 Table S5 Proteins significantly differentially expressed between the L-line and the IC-line at the restrictive temperature (29°C) Protein name MET75Cb protein precursor Glutamate oxaloacetate transaminase 2 Sarcoplasmic calcium-binding protein 1 Arginine kinase CG31883 Larval serum protein 2 Adh-related Ductus ejaculatorius peptide 99B precursor Voltage-dependent anion-selective channel Isocitrate dehydrogenase Odorant-binding protein 99b FlyBase ID FBgn0028415 trm|Q8IPY3 FBgn0020908 FBgn0000116 FBgn0032122 FBgn0002565 FBgn0000056 FBgn0250832 FBgn0004363 FBgn0001248 FBgn0039685 11 L29/IC29 1.27 0.82 0.93 0.94 0.44 0.84 1.66 1.25 1.16 0.92 1.35 P-value 8.31E-16 8.55E-12 1.19E-08 1.13E-06 1.41E-06 2.42E-05 5.37E-05 7.12E-05 7.52E-05 1.27E-04 3.31E-04 Error factor 1.05 1.05 1.03 1.03 1.35 1.08 1.20 1.10 1.07 1.04 1.16 CG8036 Mitochondrial aconitase CG15616 Esterase 6 precursor CG11151 Aldehyde dehydrogenase Accessory gland-specific peptide 70A precursor Glutamine synthetase 1 CG30415 Acyl-CoA-binding protein homolog CG3731 CG9914 Heat shock protein 60 CG15369 CG7920 Glycogen phosphorylase 6-phosphogluconate dehydrogenase, decarboxylating Protein anoxia up-regulated Glutathione S-transferase S1 CG8343 CG5844 CG6287 CG32920 CG9391 Putative ATP synthase f chain, mitochondrial 60S acidic ribosomal protein P0 Heterogeneous nuclear ribonucleoprotein at 87F 6-phosphofructo-2-kinase Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic Probable acyl-CoA dehydrogenase, mitochondrial precursor CG6045 Yippee interacting protein 2 Cytochrome c-2 FBgn0037607 FBgn0010100 FBgn0034153 FBgn0000592 FBgn0030519 FBgn0012036 FBgn0003034 FBgn0001142 FBgn0250838 FBgn0010387 FBgn0038271 FBgn0030737 FBgn0015245 FBgn0030105 FBgn0039737 FBgn0004507 FBgn0004654 FBgn0020439 FBgn0010226 FBgn0040502 FBgn0038049 FBgn0032350 FBgn0038570 FBgn0037063 FBgn0035032 FBgn0000100 FBgn0004237 FBgn0027621 FBgn0001128 FBgn0035811 FBgn0038349 FBgn0040064 FBgn0000409 12 0.93 0.94 0.87 1.29 1.11 0.93 0.89 0.84 1.21 0.86 1.11 1.11 0.89 1.22 0.93 0.96 0.89 1.56 0.94 0.70 0.69 0.88 0.92 1.17 1.18 1.22 1.27 0.89 0.95 0.90 0.91 0.93 1.09 4.86E-04 7.38E-04 9.10E-04 1.24E-03 4.53E-03 4.88E-03 6.78E-03 7.40E-03 7.72E-03 8.10E-03 8.31E-03 8.45E-03 8.88E-03 9.26E-03 9.47E-03 1.22E-02 1.24E-02 1.35E-02 1.46E-02 1.46E-02 1.53E-02 1.60E-02 1.67E-02 1.77E-02 1.91E-02 2.02E-02 2.04E-02 2.28E-02 2.50E-02 2.51E-02 2.51E-02 2.91E-02 3.12E-02 1.04 1.03 1.08 1.16 1.07 1.05 1.08 1.13 1.14 1.12 1.08 1.08 1.09 1.13 1.05 1.03 1.10 1.25 1.05 1.29 1.12 1.11 1.07 1.06 1.14 1.18 1.10 1.08 1.05 1.09 1.09 1.07 1.08 CG5162 CG10512 CG17838 CG4347 Fructose-bisphosphate aldolase CG4389 Glutathione S transferase D1 CG6439 Triose phosphate isomerase FBgn0030828 FBgn0037057 FBgn0038826 FBgn0035978 FBgn0000064 FBgn0028479 FBgn0001149 FBgn0038922 FBgn0086355 13 1.16 1.11 1.14 1.13 0.97 0.92 0.89 1.08 0.95 3.24E-02 3.24E-02 3.25E-02 3.44E-02 3.73E-02 4.44E-02 4.56E-02 4.74E-02 4.75E-02 1.14 1.10 1.12 1.12 1.03 1.09 1.12 1.08 1.05