Wildlife/Rodents/Pets as Vectors – Natural Contamination (last
advertisement

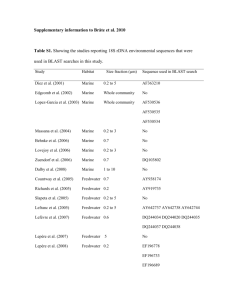
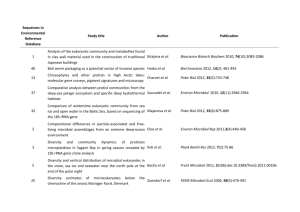
Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Pathogen E. coli O157:H7 Source wildlife Location U.S., California Prevalence and levels 1.16% of 3,202 samples positive non-O157 STEC wildlife U.S., California 7.37% of 3,202 samples positive Salmonella turtles (Trachemys scripta elegans) turtles (Emys orbicularis) Spain, eastern 15.0% of 83 samples were positive Spain, eastern 8.0% of 83 samples were positive turtles (Trachemys scripta elegans) turtles (Emys orbicularis) Spain, eastern 0.0% of 83 samples were positive Spain, eastern 0.0% of 83 samples were positive S. enterica gulls U.S., eastern shore of Virginia 17.2% of 360 fecal samples 22 serotypes and 26 PFGE DNA fingerprint patterns, including S. Newport pattern 61, were identified. E. coli O157:H7 rodents U.S., California agricultural farms 0.2% of 1,043 samples were positive Salmonella Campylobacter Campylobacter Cryptosporidium spp. S. enterica S. enterica Salmonella rodents rodents rodents amphibians U.S., California agricultural farms U.S., California agricultural farms U.S., California agricultural farms Spain Deer mice were the most abundant rodent species trapped (72.5%) 26.0% of 285 samples were positive Deer mice were the most abundant rodent species trapped (72.5%) 24.2% of 285 samples were positive Deer mice were the most abundant rodent species trapped (72.5%) 2.9% of 1,043 samples were positive Deer mice were the most abundant rodent species trapped (72.5%) 0% of 72 amphibian samples were positive Reference Cooley, M.B., M. Jay-Russell, E.R. Atwill, D. Carychao, K. Nguyen, B. Quiñones, R. Patel, S. Walker, M. Swimley, E. Pierre-Jerome, A.G. Gordus, and R.E. Mandrell. 2013. Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS One 8:e65716. Cooley, M.B., M. Jay-Russell, E.R. Atwill, D. Carychao, K. Nguyen, B. Quiñones, R. Patel, S. Walker, M. Swimley, E. Pierre-Jerome, A.G. Gordus, and R.E. Mandrell. 2013. Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS One 8:e65716. Marin, C., S. Ingresa-Capaccioni, S. González-Bodi, F. Marco-Jiménez, and S. Vega. 2013. Free-living turtles are a reservoir for Salmonella but not for Campylobacter. PLoS One 8:E72350. Marin, C., S. Ingresa-Capaccioni, S. González-Bodi, F. Marco-Jiménez, and S. Vega. 2013. Free-living turtles are a reservoir for Salmonella but not for Campylobacter. PLoS One 8:E72350. Marin, C., S. Ingresa-Capaccioni, S. González-Bodi, F. Marco-Jiménez, and S. Vega. 2013. Free-living turtles are a reservoir for Salmonella but not for Campylobacter. PLoS One 8:E72350. Marin, C., S. Ingresa-Capaccioni, S. González-Bodi, F. Marco-Jiménez, and S. Vega. 2013. Free-living turtles are a reservoir for Salmonella but not for Campylobacter. PLoS One 8:E72350. Gruszynski, K., S. Pao, C. Kim, D.M. Toney, K. Wright, A. Colón, T. Engelmeyer, and S.J. Levine. 2014. Evaluating gulls as potential vehicles of Salmonella enterica serotype Newport (JJPX01.0061) contamination of tomatoes grown on the Eastern shore of Virginia. Appl. Environ. Microbiol. 80:235-238. Kilonzo, C., X. Li, E.J. Vivas, M.T. Jay-Russell, K.L. Fernandez, E.R. Atwill. 2013. Fecal shedding of zoonotic food-borne pathogens by wild rodents in a major agricultural region of the Central California coast. Appl. Environ. Microbiol. 79:6337-6344. Kilonzo, C., X. Li, E.J. Vivas, M.T. Jay-Russell, K.L. Fernandez, E.R. Atwill. 2013. Fecal shedding of zoonotic food-borne pathogens by wild rodents in a major agricultural region of the Central California coast. Appl. Environ. Microbiol. 79:6337-6344. Kilonzo, C., X. Li, E.J. Vivas, M.T. Jay-Russell, K.L. Fernandez, E.R. Atwill. 2013. Fecal shedding of zoonotic food-borne pathogens by wild rodents in a major agricultural region of the Central California coast. Appl. Environ. Microbiol. 79:6337-6344. Kilonzo, C., X. Li, E.J. Vivas, M.T. Jay-Russell, K.L. Fernandez, E.R. Atwill. 2013. Fecal shedding of zoonotic food-borne pathogens by wild rodents in a major agricultural region of the Central California coast. Appl. Environ. Microbiol. 79:6337-6344. Briones, V., Téllez, J. Goyache, C. Ballesteros, M. del Pilar Lanzarot, L.Domínguez, and J.F. Fernández-Garayzábal. 2004. Salmonella diversity associated with wild reptiles and amphibians in Spain. Environ. Microbiol. 6:868-871. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 1 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Salmonella reptiles Spain Salmonella dog feces Italy 41.5% of 94 reptile samples were positive Subspecies Salmonella enterica enterica accounted for up to 50% of isolates. Serotypes Anatum, Herzliya, Abony, were the most frequently isolated. 0% of 418 samples were positive E. coli O157 mice, wood droppings Czech Republic 0% of 7 samples positive E. coli O157 pigeons and sparrows, domestic Czech Republic 0% of 70 samples positive E. coli O157 Rat colons Czech Republic 40% of 10 samples positive Campylobacter ducks, Mallard, wild U.K. 9.2-52.2% of fecal samples collected on five sampling occasions (60-100 wild ducks had been observed at location prior to sampling) Salmonella raccoons, feces U.S., Pennsylvania Only one isolate of the 47 serotypes of Campylobacter recovered were commonly associated with human disease. 7.4% of 738 samples were positive 7.8% of 128 samples from rural areas were positive 8.7% of 332 samples from forested areas were positive 5.7% of 278 samples from suburban areas were positive 0% of 1 sample positive Clostridium perfringens deer, feral, feces Australia, Sydney watersheds Clostridium perfringens duck, wood, feces Australia, Sydney watersheds 44.4% of 9 samples positive Clostridium perfringens fox, feral, feces Australia, Sydney watersheds 100% of 1 sample positive Clostridium perfringens goat, feral, feces Australia, Sydney watersheds 100% of 5 samples positive Clostridium perfringens kangaroo, feces Australia, Sydney watersheds 9.1% of 11 samples positive Clostridium perfringens pig, feral, feces Australia, Sydney watersheds 0% of 5 samples positive Briones, V., Téllez, J. Goyache, C. Ballesteros, M. del Pilar Lanzarot, L.Domínguez, and J.F. Fernández-Garayzábal. 2004. Salmonella diversity associated with wild reptiles and amphibians in Spain. Environ. Microbiol. 6:868-871. Cinquepalmi, V., R. Monno, L. Fumarola, G. Ventrella, C. Calia, M.F. Greco, D. de Vito, and L. Soleo. 2013. Environmental contamination by dog feces: A public health problem? Int. J. Environ. Res. Publ. Hlth. 10:72-84. Čížek, A., P. Alexa, I. Literák, J. Hamřík, P. Novák, and J. Smola. 1999. Shiga toxin-producing Escherichia coli O157 in feedlot cattle and Norwegian rats from a large-scale farm. Lett. Appl. Microbiol. 28:435439. Čížek, A., P. Alexa, I. Literák, J. Hamřík, P. Novák, and J. Smola. 1999. Shiga toxin-producing Escherichia coli O157 in feedlot cattle and Norwegian rats from a large-scale farm. Lett. Appl. Microbiol. 28:435439. Čížek, A., P. Alexa, I. Literák, J. Hamřík, P. Novák, and J. Smola. 1999. Shiga toxin-producing Escherichia coli O157 in feedlot cattle and Norwegian rats from a large-scale farm. Lett. Appl. Microbiol. 28:435439. Colles, F.M., J.S. Ali, S.K. Sheppard, N.D. McCarthy, and M.C.J. Maiden. 2011. Campylobacter populations in wild and domesticated Mallard ducks (Anas platyrhynchos). Environ. Microbiol. Rep. 3:574580. Compton, J.A., J.A. Baney, S.C. Donaldson, B.A. Houser, G.J. San Julian, R.H. Yahner, W. Chmielecki, S. Reynolds, and B.M. Jayarao. 2008. Salmonella infections in the common raccoon (Procyon lotor) in Western Pennsylvania. J. Clin. Microbiol. 46:3084-3086. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 2 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Clostridium perfringens platypus, feces Australia, Sydney watersheds 0% of 11 samples positive Clostridium perfringens possum, brushtail, feces Australia, Sydney watersheds 0% of 2 samples positive Clostridium perfringens rabbit, feral, feces Australia, Sydney watersheds 0% of 4 samples positive Clostridium perfringens rat, feces Australia, Sydney watersheds 0% of 5 samples positive Clostridium perfringens wallaby, feces Australia, Sydney watersheds 0% of 10 samples positive Clostridium perfringens wombat, feces Australia, Sydney watersheds 28.6% of 7 samples positive Cryptosporidium deer, feral, feces Australia, Sydney watersheds 100% of 1 sample positive Cryptosporidium duck, wood, feces Australia, Sydney watersheds 0% of 9 samples positive Cryptosporidium goat, feral, feces Australia, Sydney watersheds 0% of 3 samples positive Cryptosporidium kangaroo, feces Australia, Sydney watersheds 36.4% of 11 samples positive Cryptosporidium pig, feral, feces Australia, Sydney watersheds 0% of 5 samples positive Cryptosporidium platypus, feces Australia, Sydney watersheds 0% of 6 samples positive Cryptosporidium possum, brushtail, feces Australia, Sydney watersheds 100% of 2 samples positive Cryptosporidium rabbit, feral, feces Australia, Sydney watersheds 50% of 2 samples positive Cryptosporidium rat, feces Australia, Sydney watersheds 0% of 4 samples positive Cryptosporidium wallaby, feces Australia, Sydney watersheds 0% of 10 samples positive Cryptosporidium wombat, feces Australia, Sydney watersheds 0% of 5 samples positive Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 3 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Giardia deer, feral, feces Australia, Sydney watersheds 0% of 1 sample positive Giardia duck, wood, feces Australia, Sydney watersheds 44.4% of 9 samples positive Giardia fox, feral, feces Australia, Sydney watersheds 100% of 1 sample positive Giardia goat, feral, feces Australia, Sydney watersheds 0% of 3 samples positive Giardia kangaroo, feces Australia, Sydney watersheds 0% of 11 samples positive Giardia pig, feral, feces Australia, Sydney watersheds 0% of 5 samples positive Giardia platypus, feces Australia, Sydney watersheds 0% of 6 samples positive Giardia possum, brushtail, feces Australia, Sydney watersheds 0% of 2 samples positive Giardia rabbit, feral, feces Australia, Sydney watersheds 0% of 2 samples positive Giardia rat, feces Australia, Sydney watersheds 0% of 4 samples positive Giardia wallaby, feces Australia, Sydney watersheds 0% of 10 samples positive Giardia wombat, feces Australia, Sydney watersheds 0% of 5 samples positive E. coli, STEC deer, red and roe feces Germany 70% and 73% of 30 samples each were positive for red deer and roe deer, respectively. Salmonella enterica birds, feces or cloaca U.S., California 6.6% of 105 samples were positive Salmonella enterica blackbird, feces U.S., California 0% of 57 samples were positive Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Cox, P., M. Griffith, M. Angles, D. Deere, and C. Ferguson. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929-5934. Eggert, M., E. Stüber, M. Heurich, M. Fredriksson-Ahomaa, Y. Burgos, L. Beutin, and E. Märtlbauer. 2013. Detection and characterization of Shiga toxin-producing Escherichia coli in faeces and lymphatic tissue of free-ranging deer. Epidemiol. Infection. 141:251259. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 4 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Salmonella enterica coyote, feces U.S., California 5% of 40 samples were positive Salmonella enterica deer, feces U.S., California 1.9% of 104 samples were positive Salmonella enterica elk, feces U.S., California 2.6% of 39 samples were positive Salmonella enterica geese, feces U.S., California 0% of 17 samples were positive Salmonella enterica junco, feces U.S., California 0% of 2 samples were positive Salmonella enterica mallard, feces U.S., California 0% of 3 samples were positive Salmonella enterica mouse, feces U.S., California 0% of 24 samples were positive Salmonella enterica pig, wild, feces U.S., California 2.4% of 41 samples were positive Salmonella enterica rabbit, feces U.S., California 0% of 57 samples were positive Salmonella enterica raccoon, feces U.S., California 0% of 2 samples were positive Salmonella enterica skunk, feces U.S., California 30.7% of 13 samples were positive Salmonella enterica squirrel, feces U.S., California 0% of 28 samples were positive Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 5 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Salmonella enterica titmouse, feces U.S., California 0% of 1 sample was positive Salmonella enterica warbler, feces U.S., California 0% of 1 sample was positive Salmonella enterica woodpecker, feces U.S., California 0% of 1 sample was positive Cryptosporidium pigs, feral Australia, west 0.3% of 292 samples were positive Giardia pigs, feral Australia, west 1.7% of 292 samples were positive E. coli O157:H7 bird feces collected from cattle farm U.S., Northwest 0.5% of 200 pooled samples positive E. coli O157:H7 cat feces collected from cattle farm U.S., Northwest 0% of 33 samples positive E. coli O157:H7 dog feces collected from cattle farm U.S., Northwest 3.1% of 65 samples positive E. coli O157:H7 flies collected from cattle farm U.S., Northwest 3.3% of 60 pooled samples positive E. coli O157:H7 rodents collected from cattle farm U.S., Northwest 0% of 300 samples positive E. coli O157:H7 wildlife, misc. feces collected from cattle farm U.S., Northwest 0% of 34 pooled samples Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Gorski, L, C.T. Parker, A. Liang, M.B. Cooley, T.M. Jay-Russell, A.G. Gordus, E.R. Atwill, and R.E. Mandrell. 2011. Prevalence, distribution, and diversity of Salmonella enterica in a major produce region of California. Appl. Environ. Microbiol. 77:2734-2748. Hampton, J., P.B.S. Spencer, A.D. Elliot, and R.C.A. Thompson. 2006. Prevalence of zoonotic pathogens from feral pigs in major public drinking water catchments in Western Australia. EcoHealth 3:103-108. Hampton, J., P.B.S. Spencer, A.D. Elliot, and R.C.A. Thompson. 2006. Prevalence of zoonotic pathogens from feral pigs in major public drinking water catchments in Western Australia. EcoHealth 3:103-108. Hancock, D.D., T.E. Besser, D.H. Rice, E.D. Ebel, D.E. Herriott, and L.V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35:11-19. Hancock, D.D., T.E. Besser, D.H. Rice, E.D. Ebel, D.E. Herriott, and L.V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35:11-19. Hancock, D.D., T.E. Besser, D.H. Rice, E.D. Ebel, D.E. Herriott, and L.V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35:11-19. Hancock, D.D., T.E. Besser, D.H. Rice, E.D. Ebel, D.E. Herriott, and L.V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35:11-19. Hancock, D.D., T.E. Besser, D.H. Rice, E.D. Ebel, D.E. Herriott, and L.V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35:11-19. Hancock, D.D., T.E. Besser, D.H. Rice, E.D. Ebel, D.E. Herriott, and L.V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35:11-19. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 6 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Salmonella enteritidis mice and rats, feces U.S., Maine 16.2% of 715 mice and rat samples were positive for Salmonella enteritidis. On contaminated farms, 24.0% of the mice were positive whereas on clean farms, S. enteritidis was not detected in mice. Henzler, D.J. and H.M. Opitz. 1992. The role of mice in the epizootiology of Salmonella enteritidis infection on chicken layer farms. Avian Dis. 36:625-631. S. enteritidis concentrations in the feces of one mouse yielded 2.3 x 105 CFU/fecal pellet. Campylobacter wildlife, feces Netherlands S. enteritidis persisted at least for 10 months in an infected mouse population. 13.8% of 897 samples were positive Cryptosporidium parvum wildlife, feces Netherlands Of positive samples, 59.8% were recovered from corvidae and 22.4% from meadow birds and waterfowl. 0% of 897 samples were positive E. coli O157 wildlife, feces Netherlands 0.5% of 897 samples were positive Giardia lamblia wildlife, feces Netherlands 0.01% of 897 samples were positive Salmonella wildlife, feces Netherlands 0.1% of 897 samples were positive E. coli O157:H7 swine, feral, feces U.S., Central California 14.9% of 87 samples were positive Isolates from feral swine were matched to an outbreak strain. E. coli O157:H7 Campylobacter spp. Wildlife, feces swine, feral U.S., Central California U.S., California 0% of 26 samples were positive Wildlife feces included coyote (n=1), deer (n=4), dog (n=1), horse (n=2), sheep/goat (n=3), waterfowl (n=2), unknown species (n=11) and owl (n=2) 40% of 30 feral swine gastrointestinal and oral cavity specimens were positive. Juveniles had a greater prevalence (54.5%) compared to adults (31.6%) Heuvelink, A.E., J.T.M. Zwarkruis, C. Van Heerwaarden, B. Arends, V. Stortelder, and E. de Boer. 2008. Pathogenic bacteria and parasites in wildlife and surface water. Tijdschrift Voor Diergeneeskunde 133:330-335. Heuvelink, A.E., J.T.M. Zwarkruis, C. Van Heerwaarden, B. Arends, V. Stortelder, and E. de Boer. 2008. Pathogenic bacteria and parasites in wildlife and surface water. Tijdschrift Voor Diergeneeskunde 133:330-335. Heuvelink, A.E., J.T.M. Zwarkruis, C. Van Heerwaarden, B. Arends, V. Stortelder, and E. de Boer. 2008. Pathogenic bacteria and parasites in wildlife and surface water. Tijdschrift Voor Diergeneeskunde 133:330-335. Heuvelink, A.E., J.T.M. Zwarkruis, C. Van Heerwaarden, B. Arends, V. Stortelder, and E. de Boer. 2008. Pathogenic bacteria and parasites in wildlife and surface water. Tijdschrift Voor Diergeneeskunde 133:330-335. Heuvelink, A.E., J.T.M. Zwarkruis, C. Van Heerwaarden, B. Arends, V. Stortelder, and E. de Boer. 2008. Pathogenic bacteria and parasites in wildlife and surface water. Tijdschrift Voor Diergeneeskunde 133:330-335. Jay, M.T., M. Cooley, D. Carychao, G.W. Wiscomb, R.A. Sweitzer, L. Crawford-Miksza, J.A. Farrar, D.K. Lau, J. O'Connell, A. Millington, R.V. Asmundson, E.R. Atwill, and R.E. Mandrell. 2007. Escherichia coli O157:H7 in feral swine near spinach fields and cattle, Central California coast. Emerg. Infect. Dis. 13:1908-1911. Jay, M.T., M. Cooley, D. Carychao, G.W. Wiscomb, R.A. Sweitzer, L. Crawford-Miksza, J.A. Farrar, D.K. Lau, J. O'Connell, A. Millington, R.V. Asmundson, E.R. Atwill, and R.E. Mandrell. 2007. Escherichia coli O157:H7 in feral swine near spinach fields and cattle, Central California coast. Emerg. Infect. Dis. 13:1908-1911. Jay-Russell, M.T., A. Bates, L. Harden, W.G. Miller, and R.E. Mandrell. 2012. Isolation of Campylobacter from feral swine (Sus scrofa) on the ranch associated with the 2006 Escherichia coli O157:H7 spinach outbreak investigation in California. Zoonoses and Publ. Hlth. More females had a greater prevalence (42.1%) compared to males (36.4%). Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 7 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Toxoplasma gondii Toxoplasma gondii Toxoplasma gondii Toxoplasma gondii E. coli (EHEC) Campylobacter spp. mouse, house, on organic pig farms, brain and heart tissue mouse, wood, on organic pig farms, brain and heart tissue rat, common, on organic pig farms, brain and heart tissue shrew, whitetoothed, on organic pig farms, brain and heart tissue geese, Canada, feces Netherlands 6.5% of 31 samples were positive Kijlstra, A., B. Meerburg, J. Cornelissen, S. De Craeye, P. Vereijken, and E. Jongert. 2008. The role of rodents and shrews in the transmission of Toxoplasma gondii to pigs. Vet. Parasitol. 156:183-190. Netherlands 14.3% of 7 samples were positive Kijlstra, A., B. Meerburg, J. Cornelissen, S. De Craeye, P. Vereijken, and E. Jongert. 2008. The role of rodents and shrews in the transmission of Toxoplasma gondii to pigs. Vet. Parasitol. 156:183-190. Netherlands 10.3% of 39 samples were positive Kijlstra, A., B. Meerburg, J. Cornelissen, S. De Craeye, P. Vereijken, and E. Jongert. 2008. The role of rodents and shrews in the transmission of Toxoplasma gondii to pigs. Vet. Parasitol. 156:183-190. Netherlands 13.6% of 22 samples were positive Kijlstra, A., B. Meerburg, J. Cornelissen, S. De Craeye, P. Vereijken, and E. Jongert. 2008. The role of rodents and shrews in the transmission of Toxoplasma gondii to pigs. Vet. Parasitol. 156:183-190. U.S., Colorado 6.0% of 151 samples were positive gull, California Callifornia 45% of the 159 fecal samples were positive for Campylobacter at the genus level. Most campylobacters were novel and not closely related to species commonly associated with human illness. Kullas, H., M. Coles, J. Rhyan, and L. Clark. 2002. Prevalence of Escherichia coli serogroups and human virulence factors in feces of urban Canada geese (Branta canadensis). Int. J. Environ. Hlth. Res. 12:153-162. Lu, J., H. Ryu, J.W. Santo Domingo, J.F. Griffith, and N. Ashbolt. 2011. Molecular detection of Campylobacter spp. in California gull (Larus californicus) excreta. Appl. Environ. Microbiol. 77:5034-5039. STEC (Shiga toxin-producing E. coli) boars, wild Spain, northwest C. jejuni and C. lari were detected in fewer than 2% of the 159 isolates 8.4% of 262 samples were positive STEC (Shiga toxin-producing E. coli) deer, roe Spain, northwest 53% of 179 samples were positive STEC (Shiga toxin-producing E. coli) foxes Spain, northwest 1.9% of 260 samples were positive Cryptosporidium deer feces Australia 3.1% of 950 samples were positive Mora, A., C. Løpez, G. Dhabi, A.M. Løpez-Beceiro, L.E. Fidalgo, E.D. Díaz, C. Martínez-Carrasco, R. Mamani, A. Herrera, J.E. Blanco, M. Blanco, and J. Blanco. 2012. Seropathotypes, phylogroups, Stx subtypes, and intimin types of wildlife-carried, Shiga toxin-producing Escherichia coli strains with the same characteristics as humanpathogenic isolates. Appl. Environ. Microbiol. 78:2578-2585. Mora, A., C. Løpez, G. Dhabi, A.M. Løpez-Beceiro, L.E. Fidalgo, E.D. Díaz, C. Martínez-Carrasco, R. Mamani, A. Herrera, J.E. Blanco, M. Blanco, and J. Blanco. 2012. Seropathotypes, phylogroups, Stx subtypes, and intimin types of wildlife-carried, Shiga toxin-producing Escherichia coli strains with the same characteristics as humanpathogenic isolates. Appl. Environ. Microbiol. 78:2578-2585. Mora, A., C. Løpez, G. Dhabi, A.M. Løpez-Beceiro, L.E. Fidalgo, E.D. Díaz, C. Martínez-Carrasco, R. Mamani, A. Herrera, J.E. Blanco, M. Blanco, and J. Blanco. 2012. Seropathotypes, phylogroups, Stx subtypes, and intimin types of wildlife-carried, Shiga toxin-producing Escherichia coli strains with the same characteristics as humanpathogenic isolates. Appl. Environ. Microbiol. 78:2578-2585. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 8 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Cryptosporidium rabbit feces Australia 8.4% of 263 samples were positive Cryptosporidium possum, common brushtail, feces Australia 0% of 6 samples were positive Cryptosporidium kangaroo,Eastern grey, feces Australia 2.5% of 485 samples were positive Cryptosporidium water birds feces Australia 0% of 29 samples were positive Cryptosporidium fox feces Australia 4.3% of 23 samples were positive Cryptosporidium emu feces Australia 0% of 8 samples were positive Cryptosporidium dingo feces Australia 0% of 1 sample were positive Cryptosporidium cat feces Australia 0% of 4 samples were positive Cryptosporidium dog feces Australia 3.1% of 32 samples were positive Giardia deer feces Australia 2.2% of 950 samples were positive Giardia rabbit feces Australia 1.1% of 263 samples were positive Giardia possum, common brushtail, feces Australia 0% of 6 samples were positive Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 9 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Giardia kangaroo, Eastern grey, feces Australia 6.6% of 485 samples were positive Giardia water birds feces Australia 3.4% of 29 samples were positive Giardia fox feces Australia 0% of 23 samples were positive Giardia emu feces Australia 0% of 8 samples were positive Giardia dingo feces Australia 0% of 1 sample was positive Giardia cat feces Australia 0% of 4 samples were positive Giardia dog feces Australia 3.1% of 32 samples were positive Cryptosporidium canids, wild California, Central coast 22.2% of 18 samples positive Cryptosporidium cats California, Central coast 10.8% of 74 samples positive Cryptosporidium dogs California, Central coast 1.1% of 182 samples positive Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Nolan, M.J., A.R. Jex, A.V. Koehler, S.R. Haydon, M.A. Stevens, and R.B. Gasser. 2013. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in southeastern Australia. Water Res. 47:1726-1740. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 10 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Cryptosporidium felids, wild California, Central coast 0% of 11 samples positive Cryptosporidium gulls California, Central coast 0% of 145 samples positive Cryptosporidium opossums California, Central coast 25% of 68 samples positive Cryptosporidium sea otters California, Central coast 1.0% of 103 samples positive Giardia canids, wild California, Central coast 38.9% of 18 samples positive Giardia cats California, Central coast 14.9% of 74 samples positive Giardia dogs California, Central coast 8.8% of 182 samples positive Giardia felids, wild California, Central coast 18.2% of 11 samples positive Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 11 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Giardia gulls California, Central coast 2.1% of 145 samples positive Giardia opossums California, Central coast 14.7% of 68 samples positive Giardia sea otters California, Central coast 1.0% of 103 samples positive Salmonella enterica subsp. Enterica wildlife Australia Salmonella deer, whitetailed, feces U.S., Nebraska Isolates recovered from Australian mammals or reptiles could be members of either clade A or B. However, isolates from reptiles were significantly overrepresented in clade B. Of the 41 isolates recovered from reptiles, 33(80%) were members of clade B, while only 5 (33%) of the15 isolates recovered from mammals were assigned to clade B. 1% of 500 samples were positive E. coli O157:H7 boars, wild. feces Spain, southwest Serovars isolated (Litchfield, Dessau, Infantis, and Enteritidis) are known to be pathogenic to humans and animals. 3.3% of 212 samples were positive non-O157 STEC boars, wild, feces Spain, southwest 5.2% of 212 samples were positive E. coli (VTEC) rabbits, wild U.K., Norfolk 15.5% of 129 samples were positive Positive samples were only collected in summer and not in late winter Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Oates, S.C., M.A. Miller, D. Hardin, P.A. Conrad, A. Melli, D.A. Jessup, C. Dominik, A. Roug, M.T. Tinnker, and W.A. Miller. 2012. Prevalence, environmental loading, and molecular characterization of Cryptosporidium and Giardia isolates from domestic and wild animals along the Central California coast. Appl. Environ. Microbiol. 78:87628772. Parsons, S.K.. C.M. Bull, and D.M. Gordon. 2011. Substructure within Salmonella enterica subsp. Enterica isolates from Australian wildlife. Appl. Environ. Microbiol. 77:3151-3153. Renter, D.G., D.P. Gnad, J.M. Sargeant, and S.E. Hygnstrom. 2006. Prevalence and serovars of Salmonella in the feces of free-ranging white-tailed deer (Odocoileus virginianus) in Nebraska. J. Wildlife Dis. 42:699-703. Sánchez, S., R. Martínez, A. García, D. Vidal, J. Blanco, M. Blanco, J.E. Blanco, A. Mora, S. Herrera-León, A. Echeita, J.M. Alonso, and J. Rey. 2010. Detection and characterization of O157:H7 and non-O157 Shiga toxin-producing Escherichia coli in wild boars. Vet. Microbiol. 143:420-423. Sánchez, S., R. Martínez, A. García, D. Vidal, J. Blanco, M. Blanco, J.E. Blanco, A. Mora, S. Herrera-León, A. Echeita, J.M. Alonso, and J. Rey. 2010. Detection and characterization of O157:H7 and non-O157 Shiga toxin-producing Escherichia coli in wild boars. Vet. Microbiol. 143:420-423. Scaife, H.R., D. Cowan, J. Finney, S.F. Kinghorn-Perry, and B. Crook. 2006. Wild rabbits (Oryctolagus cuniculus) as potential carriers of verocytotoxin-producing Escherichia coli. Vet. Rec. 159:175-178. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 12 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) Salmonella birds, cloacal swabs Denmark 1.5% of 1285 samples were positive Salmonella was detected in birds on farms carrying Salmonella-positive production animals (more often from cattle herds than pig herds) and only during the periods when Salmonella was detected in the production animals. Salmonella dogs and cats, feces Denmark The following birds had positive samples: barn swallow (1); blackbird (8); chaffinch (1); house sparrow (4); pied wagtail (3); starling (1); Willow warbler (1); great tit (1); Significantly associated with the food preferences of the birds 6.5% of 46 samples were positive Salmonella rodents, feces Denmark 2% of 135 samples were positive Clostridium difficile pigs, feral, feces U.S., North Carolina 4.4% of 161 samples were positive Salmonella pigs, feral, feces U.S., North Carolina 5.0% of 161 samples were positive Campylobacter spp. boars, wild Switzerland 0% of 153 tonsil samples were positive 0% of 73 fecal samples were positive Listeria monocytogenes boars, wild Switzerland 26 (17%) of 153 tonsil samples were positive 1 (1%) of 73 fecal samples were positive Salmonella spp. boars, wild Switzerland L. monocytogenes strains belonged to serotypes 1/2a (42%), 1/2b (19%), and 4b (38%). 19 (12%) of 153 tonsil samples were positive 0% of 73 fecal samples were positive Most of the Salmonella spp. strains were of serotype Salmonella Enteritidis (75%) followed by serotypes Salmonella Stourbridge (13%) and Salmonella Veneziana (13%). 14 (9%) of 153 tonsil samples were positive 0% of 73 fecal samples were positive stx-positive Escherichia coli boars, wild Switzerland Yersinia enterocolitica boars, wild Switzerland 53 (35%) of 153 tonsil samples were positive 4 (5%) of 73 fecal samples were positive Yersinia pseudotuberculosis boars, wild Switzerland 30 (20%) of 153 tonsil samples were positive 0% of 73 fecal samples were positive Skov, M.N., J.J. Madsen, C. Rahbek, J. Lodal, J.B. Jespersen, J.C. Jørgensen, H.H. Dietz, M. Chriél, and D.L. Baggesen. 2008. Transmission of Salmonella between wildlife and meat-production animals in Denmark. J. Appl. Microbiol. 105:1558-1568. Skov, M.N., J.J. Madsen, C. Rahbek, J. Lodal, J.B. Jespersen, J.C. Jørgensen, H.H. Dietz, M. Chriél, and D.L. Baggesen. 2008. Transmission of Salmonella between wildlife and meat-production animals in Denmark. J. Appl. Microbiol. 105:1558-1568. Skov, M.N., J.J. Madsen, C. Rahbek, J. Lodal, J.B. Jespersen, J.C. Jørgensen, H.H. Dietz, M. Chriél, and D.L. Baggesen. 2008. Transmission of Salmonella between wildlife and meat-production animals in Denmark. J. Appl. Microbiol. 105:1558-1568. Thakur, S., M. Sandfoss, S. Kennedy-Stoskopf, and C.S. DePerno. 2011. Detection of Clostridium difficile and Salmonella in feral swine population in North Carolina. J. Wildlife Dis. 47:774-776. Thakur, S., M. Sandfoss, S. Kennedy-Stoskopf, and C.S. DePerno. 2011. Detection of Clostridium difficile and Salmonella in feral swine population in North Carolina. J. Wildlife Dis. 47:774-776. Wacheck, S., M. Fredriksson-Ahomaa, M. Konig, A. Stolle, and R. Stephen. 2010. Wild boars as an important reservoir for foodborne pathogens. Foodborne Path. Dis. 7:307-312. Wacheck, S., M. Fredriksson-Ahomaa, M. Konig, A. Stolle, and R. Stephen. 2010. Wild boars as an important reservoir for foodborne pathogens. Foodborne Path. Dis. 7:307-312. Wacheck, S., M. Fredriksson-Ahomaa, M. Konig, A. Stolle, and R. Stephen. 2010. Wild boars as an important reservoir for foodborne pathogens. Foodborne Path. Dis. 7:307-312. Wacheck, S., M. Fredriksson-Ahomaa, M. Konig, A. Stolle, and R. Stephen. 2010. Wild boars as an important reservoir for foodborne pathogens. Foodborne Path. Dis. 7:307-312. Wacheck, S., M. Fredriksson-Ahomaa, M. Konig, A. Stolle, and R. Stephen. 2010. Wild boars as an important reservoir for foodborne pathogens. Foodborne Path. Dis. 7:307-312. Wacheck, S., M. Fredriksson-Ahomaa, M. Konig, A. Stolle, and R. Stephen. 2010. Wild boars as an important reservoir for foodborne pathogens. Foodborne Path. Dis. 7:307-312. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 13 Wildlife/Rodents/Pets as Vectors – Natural Contamination (last updated 1/12/2014) E. coli O157:H7 European starlings U.S., Ohio 1.2% of 430 starling intestinal samples were positive Williams, M.L., D.L. Pearl, and J.T. LeJeune. 2011. Multiple-locus variable nucleotide tandem repeat subtype analysis implicates European starlings as biological vectors for Escherichia coli O157:H7 in Ohio, USA. J. Appl. Microbiol. 111:982-988. Compiled by Marilyn Erickson, Center for Food Safety, University of Georgia Downloaded from the website: A Systems Approach for Produce Safety: A Research Project Addressing Leafy Greens found at: http://www.ugacfs.org/producesafety/index.html. See http://www.ugacfs.org/producesafety/Pages/TermsofUse.html for disclaimers & terms for use of information in this document. Page 14