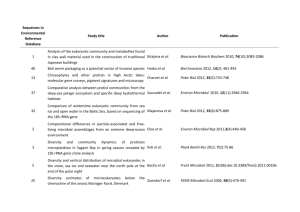
Selected studies describing the design and application of Functional
advertisement

Selected studies describing the design and application of Functional Gene Arrays: Gao, H., Z. K. Yang, T. J. Gentry, L. Wu, C. W. Schadt, and J. Zhou. 2007. Microarray-based Analysis of Microbial Community RNAs by Whole Community RNA Amplification (WCRA). Appl. Environ. Microbiol 73: 563-571 . He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, Wu WM, Gu B, Jardine P, Criddle CS, and J-Z Zhou. 2007. GeoChip: A novel comprehensive microarray for investigating biogeochemical and environmental processes. ISME Journal 1:67-77 Li, X., Z. He, and J. Zhou. 2005. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucl. Acids Res. 33:61146123. Liebich, J., C. W. Schadt, S. C. Chong, Z. L. He, S. K. Rhee, and J. Z. Zhou. 2006. Improvement of oligonucleotide probe design criteria for functional gene microarrays in environmental applications. Appl. Environ. Microbiol.72:1688-1691. Schadt, C., and J. Zhou. 2006. Advances in Microarray-Based Technologies for Soil Microbial Community Analyses pp189-203. in Soil Biology, vol. 8., Springer-Verlag. Schadt, C. W., J. Liebich, S. C. Chong, T. J. Gentry, Z. L. He, H. B. Pan, and J. Z. Zhou. 2005. Design and use of functional gene microarrays (FGAs) for the characterization of microbial communities, pp331-355, in Microbial Imaging, vol. 34., Elsevier. Wu, L., X. Liu, C. W. Schadt, and J. Zhou. 2006. Microarray-Based Analysis of Subnanogram Quantities of Microbial Community DNAs by Using Whole-Community Genome Amplification. Appl. Environ. Microbiol. 72:4931-4941. Zhou, J-Z., S. Kang, C.W. Schadt, and C.T. Garten. 2008. Spatial scaling of functional gene diversity across various microbial taxa. PNAS 105:7768-7773 Functional Gene Markers in Fungal Ecology: Blackwood, C. B., M. P. Waldrop, D. R. Zak, and R. L. Sinsabaugh. 2007. Molecular analysis of fungal communities and laccase genes in decomposing litter reveals differences among forest types but no impact of nitrogen deposition. Environ. Microbiol. 9:1306–1316. Edwards I.P., Upchurch R.A. and D.R. Zak. 2008. Isolation of Fungal Cellobiohydrolase I genes from sporocarps and forest soils by PCR. Appl. Environ. Microbiol 74:3481-3489. Luis, P., G. Walther, H. Kellner, F. Martin, and F. Buscot. 2004. Diversity of laccase genes from basidiomycetes in a forest soil. Soil. Biol. Biochem. 36:1025–1036. Lyons, J. I., S. Y. Newell, A. Buchan, and M. A. Moran. 2003. Diversity of ascomycete laccase gene sequences in southeastern US salt marsh. Microb. Ecol. 45:270–281. Reviews of Functional Genes in Microbial Ecology: Philippot, L. and S. Hanlin. 2005. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opinion. Microbiol. 8:234-239. Ward, B.B. 2005. Molecular approaches to marine microbial ecology and the marine nitrogen cycle. Annual Review of Earth and Planetary Sciences 33: 301 Zher, J.P., B.D. Jenkins, S.M. Short and G.F. Steward. 2003. Minireview: Nitrogenase gene diversity and microbial community stucture: A cross-system comparison. Environ. Microbiol. 5:539-554.