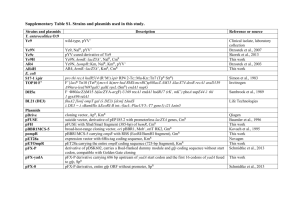
Liang et al. – Supplementary Table – 1 Supplementary Table 1
advertisement

Liang et al. – Supplementary Table – 1 Supplementary Table 1 Strains and plasmids used in this study Strains or plasmid Genotype or relevant characteristics Source or reference Escherichia coli supE44, ΔlacU169(Φ80lacZΔM15), hsdR17, DH5α recA1, gyrA96, thi-1, relA1 Agrobacterium a Kmr derivative of EHA101, Rifr Hood et al. (1990) Vector containing the P1 phage Cre site-specific Sauer and Henderson (1990) Invitrogen tumefaciens EHA105 pBS185 recombinase gene, Ampr pGEM-T Cloning vector for PCR products, Amp r Promega pGEM-TCre P1 phage Cre site-specific recombinase This study PCR-amplified from pBS185 and cloned into pGEM-T , Ampr pRT104 Vector containing the CaMV 35S promoter and a Töpfer et al. (1987) poly-(A) signal, Ampr pBluescript II SK High copy number ColE1-based phagemid, Ampr Stratagene pBluescript-loxP pBluescript II SK derivative containing two loxP This study sites pBI101 Cloning vector containing the β-glucuronidase Jefferson et al. (1987) (Gus) gene of E. coli and the nopaline synthase gene terminator (Tnos) from pBIN19, Kmr pBluescript-loxP-Tno pBluescript II SK derivative containing a 2 kb s-loxP-Gus loxP-Tnos-loxP-Gus cassette pGreen0000 Binary vector for Agrobacterium-mediated plant This study Hellens et al. (2000) Liang et al. – Supplementary Table – 2 transformation, requires the pSoup helper plasmid for replication in A. tumefaciens, Kmr pSoup Helper plasmid required for replication of Hellens et al. (2000) pGreen0000 in A. tumefaciens , Kmr pGreen-loxP pGreen0000 derivative carrying a 2.7 kb This study 35S-loxP-Tnos-loxP-Gus cassette (Fig. 1B), Kmr pGreen-Cre pGreen0000 derivative carrying a 1.5-kb 35S-Cre This study sequence (Fig.1A), Kmr pCambia2301 Binary vector for Agrobacterium-mediated plant http://www.cambia.org transformation, Kmr pCambia-loxP pCambia2301 derivative carrying a 2.7-kb This study 35S-loxP-Tnos-loxP-Gus cassette (Fig. 1B), Kmr pCambia-Cre pCambia2301 derivative carrying a 1.5-kb This study 35S-Cre sequence (Fig. 1A), Kmr pPZP112 Binary vector for Agrobacterium-mediated plant Hajdukiewicz et al. (1994) transformation, Cmr pPZP-loxP pPZP112 derivative carrying a 2.7-kb This study 35S-loxP-Tnos-loxP-Gus cassette (Fig. 1B), Cmr pISV2678 Binary vector for Agrobacterium-mediated plant M. Schultze and transformation; derivative of pGPTV-BAR E. Kondorosi CNRS, (Becker et al. 1992) with a double CaMV 35S Gif-sur-Yvette, France promoter and a translational enhancer sequence from pBI-426 (Datla et al. 1991), Kmr pISV-Cre pISV2678 derivative carrying a 1.5-kb 35S-Cre This study sequence (Fig.1A), Kmr Rifr, Ampr, Kmr, Cmr, resistance to rifampin, ampicillin, kanamycin and chloramphenicol, respectively. Liang et al. – Supplementary Table – 3 References Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20:1195-1197 Datla RS, Hammerlindl JK, Pelcher LE, Crosby WL, Selvaraj G (1991) A bifunctional fusion between beta-glucuronidase and neomycin phosphotransferase: a broad-spectrum marker enzyme for plants. Gene 101:239-246 Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989-994 Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42:819-832 Hood EE, Clapham DH, Ekberg I, Johannson T (1990) T-DNA presence and opine production in tumors of Picea abies (L.) Karst induced by Agrobacterium tumefaciens A281. Plant Mol Biol 14:111-117 Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901-3907 Sauer B, Henderson N (1990) Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol 2:441-449 Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH (1987) A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res 15:5890