A Case Study in Using Local IO and GPFS to Improve Simulation
advertisement

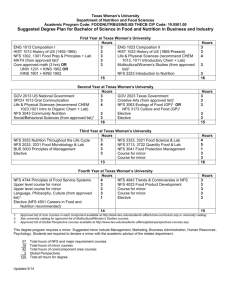
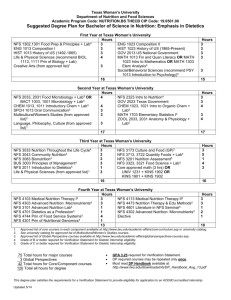
A Case Study in Using Local I/O and GPFS to Improve Simulation Scalability Vincent Bannister, G.W. Howell, C. T. Kelley, Eric Sills, Qianyi Zhang North Carolina State University1 1Contact Information: Vince Bannister, now at Microsoft, e-mail vebannis@ncsu.edu, Gary Howell and Eric Sills, HPC/ITD NCSU, Tim Kelley from the NCSU Math Department, Qianyi Zhang from the NCSU Statistics Department gary_howell@ncsu.edu, eric_sills@ncsu.edu, ctk@ncsu.edu, qzhang3@stat.unity.ncsu Abstract Many optimization algorithms exploit parallelism by calling multiple independent instances of the function to be minimized, and these function in turn may call off-the-shelf simulators. The I/O load from the simulators can cause problems for an NFS file system. In this paper we explore efficient parallelization in a parallel program for which each processor makes serial calls to a MODFLOW simulator. Each MODFLOW simulation reads input files and produces output files. The application is "embarassingly" parallel except for disk I/O. Substitution of local scratch as opposed to global file storage ameliorates synchronization and contention issues. An easier solution was to use the high performance global file system GPFS instead of NFS. Compared to using local I/O, using a high performance shared file system such as GPFS requires less user effort. Keywords: Software, Efficiency, Scalability, Parallel, High Performance File Systems, GPFS. Introduction MODFLOW is a large serial simulation suite. A parallel recode would not be easy. Due to the complexity of MODFLOW, we preferred to consider it as a “black box”. Given an input file, MODFLOW produces a desired output. Each processor can run many such simulations, with each MODFLOW output parsed by a high level code (Matlab in our case) which writes a file with the correct inputs for the next run. On a single CPU, file I/O requires only a small fraction of execution time. Parallel execution with many instances of MODFLOW and Matlab simultaneously accessing a single NFS file system produces an I/O bound problem, with file system contention resulting in poor scaling. We solved the problem in two ways, first by using local hard drives (each node has a local hard drive) for intermediate computations, second by using the high performance shared parallel file system GPFS. Both solutions gave good performance for a moderate number of processors. This paper describes and compares the solutions. First the problem is described in a bit more detail, then the steps taken to use local file systems as opposed to a global file system. Timing results are compared to the solution of changing the file system from NFS to GPFS. Discussion and conclusions are in a final section. A SAS based statistical analysis comparing sets of times for NFS, vs. NFS with local file I/O, and GPFS file systems. For each of these 3 cases we compare performance of loaded and unloaded file global file systems. A Case Study in Simulation -- Using Local I/O Our simulations used MODFLOW[1]. The MODFLOW[2] simulator creates and tears down a large number of scratch files during execution, primarily as intermediaries in the computational process. Unfortunately, without a major recode, it seems impossible to avoid use of the file system. The initial input is a large matrix of input values, passed in plaintext form in a single file. The program then distributes the data to each individual instance of the simulator, invoking individual simulations which produce individual output on each processor. A high level wrapper program (written in Matlab) makes decisions for a new simulator run. After a set of simulations is performed on each processor, then a final combined data file is formed. More specifically, each instance of the wrapper program first parses the entirety of the input file and forms a series of directories, numbered according to relevant job. The files relevant to the running of the simulator are then copied into each numbered directory, and each distinct input set is written to the appropriate directory. The simulator is executes via a blocking Unix system call and, after MODFLOW’s completion, the wrapper parses the appropriate output file, and removes the temporary files associated with the run. The wrapper then checks for further jobs, and if it finds there are none, synchronizes with the remaining processes to assemble the output array, and delete the remains of the directories. Once the output array is formed, the wrapper writes a common output data file in human-legible format. This approach depends only on the availability of Unix-style system commands, and implementation of the generic MPI functions. This method requires little communication overhead; each simulation is run as a serial command and the "scratch" file it writes can be read by a new simulation on the same processor. Limiting use of the shared file system to initiation of the simulation and constructing a common data file allows effective parallel scaling of the computation. File System Race Conditions in NFS The parallel code is a wrapper on the serial simulations. The wrapper initializes and sequences the serial simulations, each of which reads from an input file and creates an output file to be read by another simulation. The initial wrapper suffered from erratic NFS file system behavior, delivering only partially correct results and with some simulations aborting with input errors. The problem was that C file system calls could encounter race conditions. Though a file has been closed (or even in some cases been subjected to an "fsync"), the call will return before the file has been written to disk. This can result in a race condition between the file write performed by the simulator and the file read of the wrapper, as the write command may not have completed it’s action when the next wrapper command (read) is invoked. A pathological example: … system(“Run the simulation suite”); fp = fopen(“output file”, "a+"); … The read will then open and lock the file, forcing the write to hold, and causing the reading of an empty file, pushing null values into the results matrix. Afterward, the write will perform, generating the output as appropriate, and leaving no indication of having been interrupted. If there are a number of commands between the system call to the simulator and the read action of the wrapper, the code tends to execute correctly. However, when the two I/O calls are adjacent as in the example, the failure emerged frequently, causing as many as a fourth of the run attempts to fail, or return partial results, with filling zero or null values. To resolve this issue, the processes must be forced to block until the final command of the simulator has committed its action upon the file. On our NFS file system the standard "fsync" command did not reliably enforce synchronization. The following sleep(1) command did eliminate the race condition. One solution: … system(“Run the simulation suite”); sleep(1); fp = fopen(“output file”, "a+"); … Contention for a Shared (Shared) File System As the environment we operate is shared with other processes and users, the node-to-storage link is in near continual use. Moreover, simulations on each processor of the parallel job tend to access the shared file system simultaneously. Each process would have to frequently wait a significant amount of time to gain access to the file system. Parallel speed-ups and scalability were limited and erratic. At low network usage on NFS share: 60 Number of processors 50 40 30 20 10 0 0 2 4 6 8 10 Time for full code excecution 12 14 16 18 Use of On Node (Local) Storage The NC State blade center has 40 Gbyte local disks on each 2 CPU blade. Most of the local hard drives is available as a /scratch directory accessible to user level programs. running cluster has fairly large on-node storage, and only two processors per node. The wrapper code was modified so that intermediate simulations run to and from local storage. In the modified version, the wrapper created the individual run directories on the blades directly, ran the simulator, cleaned the blade’s temporary files, and returned the results, through MPI communication. With these changes, even during an extremely high-use period, the processes did not slow noticeably once admitted to the running queue, and performed more consistently across numerous comparable runs. As an example of a similar use of on-node storage, see Saltz et. al[3]. The code change was Essentially from this: … system(“Run the simulation suite”); fp = fopen(“output file”, "a+"); … To this: … system("mkdir /scratch/sim"); system("cp lib/* /scratch/sim/"); chdir("/scratch/sim/"); system(“Run the simulation suite”); fp = fopen(“output file”, "a+"); system(“rm –f /scratch/sim/*”); … The following results were typical for NFS results on a relatively unloaded file system. 60 Number of processors 50 40 On node FS 30 Shared FS 20 10 0 0 5 10 Time in seconds 15 20 Good parallel efficiency resulted. A disadvantage to this approach is that local file systems may not be as reliable (for example, they may be already full, or the user code may leave them full, rendering them unusable for the next run, or user). Using a high performance parallel file system (GPFS as opposed to NFS) was an alternative approach we evaluated. GPFS GPFS is a licensed commerical file system available from IBM. GPFS[4] is designed for high throughput and also to cope well with contention for the disk resource. For the following results, both GPFS and NFS file systems used similar Apple X-Raid boxes. The NFS was mounted with meta data cached. The "shared" times had all files read to and from a shared NFS file system and did the "GPFS" runs. The "On Node" runs used the NFS file system for initial and final storage of data and used local on node hard drives for intermediate input output scratch space for the MODFLOW simulations. As with the previous results, times were averaged over 50 runs, for which the 3 longest and shortest times were omitted. Number of ProcessorsOn node GPFS 16 12 8 6 4 2 1 6.71 8.46 9.69 10.3 13.26 18.9 27.2 Shared 7.91 10.96 12.96 16.1 20.7 35.7 59.3 8.49 11.03 12.18 16.2 18.6 30.6 49.3 Impact of File System Load on Execution Time The above results did not differentiate between an unloaded and a loaded file system. As another experiment, we ran the same code with 16 processes running. There were 3 factors to be tested NFS vs. GPFS, on node scratch space vs. Shared file system, and loaded file system vs. Unloaded. Both the GPFS and NFS file systems were running on the same disk hardware (Apple Xraids). For these experiments the NFS file system was used only by the test code. The GPFS file system was shared by other users, so was not as reliably unloaded. The program bonnie++ which tests I/O latency and throughput was applied to load both the NFS and GPFS file systems, with local I/O to blade hard The following 20 times for each case were on successive runs with a fresh copy of bonnie started before the first run and continuing during all 20 runs. Times from the first few minutes of the runs correspond to bonnie writing with "putc". The "putc" bonnie writes appear to be the most disruptive of the bonnie file operations. GPFS was relatively unimpacted by the running of bonnie, so we retested running 4 simultaneous versions of bonnie, verifying that bonnie was having some influence. Running from NFS file system /amd64_share bonnie on, times for successive runs (max = 134.4, median = 27.5) 50.0 62.0 41.0 38.1 35.3 76.3 134.4 27.0 27.6 27.4 26.7 27.5 27.2 27.9 27.6 28.2 25.1 26.0 30.6 25.6 Running from NFS file system /amd64_share, bonnie off, times for successive runs (max = 26.4, median = 25.1) 26.4 25.1 24.9 24.9 25.0 25.6 25.0 25.1 24.8 25.2 25.0 25.1 25.0 24.9 24.9 25.3 25.3 24.8 25.1 25.2 Running from NFS file system /amd64_share, intermediate I/O to local disks, bonnie on (max = 48.4, median = 23.8) 23.9 23.5 23.9 24.1 23.7 23.5 23.8 23.7 24.1 44.4 48.4 23.5 32.9 28.4 23.8 24.1 23.8 27.6 24.0 23.8 Running from NFS file system /amd64_share, intermediate I/O to local disks, bonnie off (max = 24.3, median = 23.7) 24.0 23.7 24.3 24.2 23.8 23.7 24.1 23.7 23.7 23.6 23.9 23.7 23.7 23.7 23.6 23.7 23.9 23.4 23.6 23.7 Running from /gpfs_share, bonnie on (max = 24.1, median = 23.6) 24.4 23.9 23.5 23.8 23.4 23.4 24.0 23.4 23.4 24.0 23.3 23.6 23.8 23.2 23.5 23.5 23.4 23.4 23.7 23.4 Running from /gpfs_share, bonnie off (max = 24.6, median = 23.7) 24.6 23.6 23.6 23.8 23.8 23.6 23.7 23.6 23.7 23.7 23.8 23.6 23.6 23.4 23.5 23.9 23.6 23.4 23.7 23.4 Running from gpfs_share, 4 copies of bonnie on (max = 29.9, median = 27.4) 29.9 27.8 27.4 28.0 27.2 27.4 27.8 27.3 27.3 27.2 27.8 27.4 27.5 27.1 27.3 27.3 27.3 27.8 27.8 27.4 29.10 Discussion A statistical analysis using SAS is appended. It shows that the assumption that the above 6 lots of 20 numbers each are drawn from the same normally distributed set of numbers is very low. An interested reader can refer to that analysis for more detail. Some conclusions supported by the data analysis are: For both an unloaded and a loaded file system, GPFS execution times were much less variable than the NFS times. When the file system was loaded, NFS times were unpredictable and especially in the case that NFS was used to store intermediate results NFS times were sometimes very poor. Putting intermediate results on local file systems improved the worst case performance of NFS but a few runs still took almost twice as long as the average. Using a global NFS file system for all the runs in the presence of file system contention (here artificially induced by running the I/O transfer test program bonnie++) caused a number of the simulations to require much more time. One run required more than 2 minutes, compared to the median execution time of around 27 seconds. Using local file ystems for scratch storage improved execution times, both in the median and maximal observed times. With bonnie on, the maximal time was 48.4 seconds, the median time was less than 24 seconds, with two observed times more than 40 seconds, and one more than 30 seconds. Without bonnie, the median execution time changed little, but the maximal time was 24.3 seconds. The variability of execution times was much less with GPFS. The maximal and median execution times were almost the same for the unloaded NFS storage with local scratch and for the GPFS without scratch and even for the GPFS with bonnie contention. Running four copies of bonnie simultaneously verified that bonnie was producing a gpfs load, but even in that case the maximal time increased by only about 20% and the median time by about 10%. In the experiments here, GPFS is faster than NFS for the case that several processes write to and read from the same file; a design difference that may explain the difference in performance is that GPFS uses distributed locking and meta data mechanisms to allow several processes to access a file simultaneously. GPFS suffers less contention due to multiple processes simultaneously accessing their own files than does a globally shared NFS, perhaps because GPFS is designed to write to multiple disks in parallel. MPI I/O compatibility is built into GPFS; configuring NFS to work with MPI I/O, on the other hand, entailed turning off caching of meta data, which had a strongly adverse impact on data throughput (the runs made here used cached meta data). For more detail about GPFS design, see[5]. While one test case of an untuned GPFS compared to an untuned (meta data cached ext3) NFS system can not be taken as definitive, GPFS performance was certainly superior in this case. We have had other user feedback indicating that codes scale better on this particular GPFS file system compared to our NFS ext3 system. From the system point of view, setting up GPFS was a significant labor, but then helped all users. Training a user to use local storage helps a user at a time and entails significant user effort. Moreover, the user must also be persuaded to clean up after himself so that local hard drives do not fill with abandoned user files. Some users of local disks find a significant staging time to recover their data after each run. Other issues with local disks Even when users do not use disks local to a cluster node, some local disk system management issues remain, e.g., choice of files to permanently store on local hard drives. For example, operating system files and system commands likely to be accessed during execution must be present, and similarly shared libraries may need to be found on all nodes so they can be accessed during code execution. Storing other frequently accessed executables on local hard drives can avoid contention for a globally shared file system. Conclusions On our cluster of GiGE connected IBM blades, currently with around 500 hundred processors and several dozen jobs running at a time, GPFS performs significantly better than NFS. Using GPFS reduced file I/O variability more than using local hard drives. Shifting the algorithm to using GPFS is usually trivial from the user point of view. Installing GPFS was not trivial from a system administration standpoint and requires more file I/O servers than does NFS. The cost of licensing GPFS may be significant. Some other high performance file systems are available. These include XFS (from SGI[6]) which has a good track record on large HPC machines and is now available in an open source Linux version, the file system Lustre [7], less mature perhaps but with a few years of use at some DOE HPC sites, available in both supported and open source versions, and the open source PVFS [8]. The bibliography gives some references on hiding latency[9] and modeling performance [10] of parallel I/O and high performance file systems. Some more general references on parallel I/O are Hubovsky's thesis [11] and Sloan's recent book on Linux clusters[12]. Acknowledgements Tim Kelley and Vince Bannister acknowledge support from NSF grant DMS-04-4537 and ARO grant W911NF-06-1-0096. G. W. Howell and Qianyi Zhang acknowledge support from the NIH Molecular Libraries Roadmap for Medical Research, Grant 1 P20 HG003900-01. The cluster was partially supported by ARO grant W911NF-04-1-0276. Bibliography 1 M. G. McDonald and A. W. Harbaugh, "A Modular Three Dimensional Finite Difference Groundwater Flow Model", U.S. Geological Survey Techniques of Water Resource Investigations, Book 6, Chapter AL, Reston, VA., 1988 2 ......................................................................... MODFLO W and some related software, http://water.usgs.gov/nrp/gwsoftware/modflow.html 3 ......................................................................... Joel Saltz, Anurag Acharya, Alan Sussman, Jeff Hollingsworth, and Michael B, "Tuning the I/O Performance of Earth Science Applications", NASA Goddard http://www.cs.umd.edu/projects/hpsl/hpio/sio/ccsf_oct95.txt, 1995 4 ......................................................................... "An Introduction to GPFS", IBM white paper, http://www03.ibm.com/systems/clusters/software/whitepapers/gpfs_intro. pdf, 2006 5 ......................................................................... Frank Schmuck and Roger Haskin, "GPFS, A Shared-Disk File System for Large Computing Clusters", Proceedings of the Conference on File and Storage Technologies (FAST '02) January, 2002, pp. 231-244. 6 ......................................................................... Philip Traubman, "Scalability and Performance in Modern File Systems", SGI white paper, http://www.sgi.com/pdfs/2668.pdf 7 ......................................................................... Lustre: "A Scalable, High-Performance File System", Cluster File Systems white paper, http://www.lustre.org/docs/whitepaper.pdf 8 ......................................................................... Parallel Virtual File System, Version 2, User Guide http://www.pvfs.org/pvfs2/pvfs2-guide.html, 2003 9 ......................................................................... Xiaosong Ma, Flexible and Efficient I/O Optimization for Parallel Applications, http://moss.csc.ncsu.edu/~mueller/seminar/spring03/ma.ppt, NC State CSC seminar, spring 2003. 10 ....................................................................... Yijian Wang and David Kaeli, "Modeling and Acceleration of FileI/O Dominated Parallel Workloads", www.ece.neu.edu/students/yiwang/Analogic.ppt, Northeastern University presentation to Analogic Corporation, 2005 11 ....................................................................... Rainer Hubovsky, "Dealing with Massive Data: From Parallel I/O to Grid I/O", http://www.cs.dartmouth.edu/pario/hubovsky_dictionary.pdf, Thesis, University of Vienna, 2003. 12........................................................................Joseph P. Sloan, High Performance Linux Clusters with OSCAR, Rocks, openMosix & MPI, O'Reilly Press, 2005. Data Description: Appendix -- Statistical analysis We had six sets of 20 times each. These were the times to perform the same sets of reads writes under the following specified conditions: s : loaded NFS global file system, not using local scratch 1 s2 : s3 s4 s5 s6 s7 unloaded NFS global file system, not using local scratch : loaded NFS global file system, using local scratch : unloaded NFS global file system, using local scratch : loaded GPFS global file system, not using local scratch : unloaded GPFS global file system, not using local scratch : 4x load on GPFS global file system, not using local scratch 1 Test the difference time between NFS and GPFS Only data set of s which is for GPFS is 4x loaded. To avoid the complication, the analysis for NFS and GPFS is just based on data set s to s and the following model. 7 1 Model (1): 6 Yt D1 D2 D3 D12 D23 t X where Y ( response variable): run time : mean time when D D D D D 0 D : dummy variable which is defined as t 1 1 2 3 12 23 , 1, D1 0, D2 : NFS GPFS dummy variable which is defined as 1, D2 0, D3 : loaded unloaded dummy variable which is defined as 1, local scrach D3 non local scrach 0, interaction effect between D and D which is equal to D D D : interaction effect between D and D which is equal to D D : error which is assumed to be white noise. D12 : 1 2 23 2 3 1 2 2 3 t Note that the number of observations for NFS is 80 and the number of observations for GPFS is 40. Therefore the data is unbalanced and ANOVA table is not appropriate for the analysis of NFS and GPFS. Tukey-Kramer test, which can solve the unbalanced problem for comparison, is used to test the difference between GPFS and NFS. Following provides the SAS output for Tukey-Kramer test on the variable D . From 1 the output, it is obvious that the difference between NFS and GPFS is significant. SAS output for the means comparison 1 Test the difference between using local scratch and not using local scratch Model (1) can still be used as a regression model to test whether the difference between using local scratch and not using local scratch is significant or not. The analysis is based on data set s to s . The number of observations for using local scratch is 40 and the number of observation for not using local scratch is 80. So Tukey-Kramer test is used again for this comparison. The SAS output shows that the difference between them is significant. 1 6 SAS output for the means comparison 1 Test difference between loaded and unloaded FS. From the ANOVA table, the p-value for load ( D ) is .5344, which is not small. However, the p-value for interaction effect ( D ) in the same ANOVA table is low. Therefore, the analysis 2 12 for difference between loaded and unloaded should extend to a multiple comparison. The elements for multiple comparisons are: loaded NFS loaded GPFS unloaded NFS unloaded GPFS The output from SAS shows: No significant difference between unloaded GPFS and unloaded NFS No significant difference between unloaded GPFS and loaded GPFS No significant difference between unloaded NFS and loaded GPFS significant difference between loaded NFS and unloaded GPFS significant difference between loaded NFS and loaded GPFS significant difference between loaded NFS and unloaded NPFS Therefore, the above four elements can be view as two groups: Group 1: loaded NFS Group 2: unloaded GPFS, loaded GPFS and unloaded NFS There is significant difference between groups. SAS output for ANOVA Table and means comparison: 1 Test for the effect of load in GPFS. From the data description, there are three levels for load in GPFS. To test the effect of load in GPFS, make analysis based on data set s to s and use the following regression model. 5 Model (2): 7 Yt D D 2 23 t , where Y ( response variable): run time 0 : mean time when D D : classified variable which is defined as t D2 2 2 0, unloaded D2 1, one loaded 2, four loaded D3 : dummy variable which is defined as 1, local scrach D3 non local scrach 0, D2 D3 interaction effect between and which is equal to D D : error which is assumed to be white noise. D23 : 2 t The multiple comparison results from SAS shows: No significant difference between unloaded and one loaded Significant difference between one loaded and four loaded 3 Significant difference between unloaded and four loaded The GLM Procedure Least Squares Means load LSMEAN time LSMEAN 0 1 2 23.6800000 23.6000000 27.6000000 Number 1 2 3 Least Squares Means for effect load Pr > |t| for H0: LSMean(i)=LSMean(j) Dependent Variable: time i/j 1 2 3 The GLM Procedure Dependent Variable: time Source Mean Square Model 777.85588 Sum of DF Squares F Value Pr > F 6.37 Error 122.14725 5 <.0001 3889.27942 114 13924.78650 Corrected Total MSE R-Square time Mean 119 17814.06592 Coeff Var Root The GLM Procedure Least Squares Means Adjustment for Multiple Comparisons: Tukey-Kramer H0:LSMean1= local LSMean2 time LSMEAN Pr 0 27.9962500 1 21.0087500 > |t| 0.0055 The GLM Procedure Least Squares Means Adjustment for Multiple Comparisons: Tukey-Kramer NFS H0:LSMean1= LSMean2 time LSMEAN Pr > |t| 0 20.1462500 1 28.8587500 0.0006 1 Test the difference between s and s Use the two sample t-test to test whether the mean of s is different from the mean of s . The SAS output for the test is 3 5 3 5 The TTEST Procedure Statistics Lower CL Upper CL Lower CL Upper CL Variable D1 N Mean Mean Mean Std Dev Std Dev Std Dev Std Err time 0.2275 time 5.3795 time 4.0915 0 20 23.46 23.6 23.74 0.2991 0.4369 0.0669 1 20 23.634 26.945 30.256 7.0738 10.332 1.5817 Diff (1-2) -6.55 -3.345 -0.14 5.0064 6.4521 1.5832 T-Tests DF Variable Method t Value Pr > |t| Variances The means of the two samples are significantly different from each other.