Male genital organs
advertisement
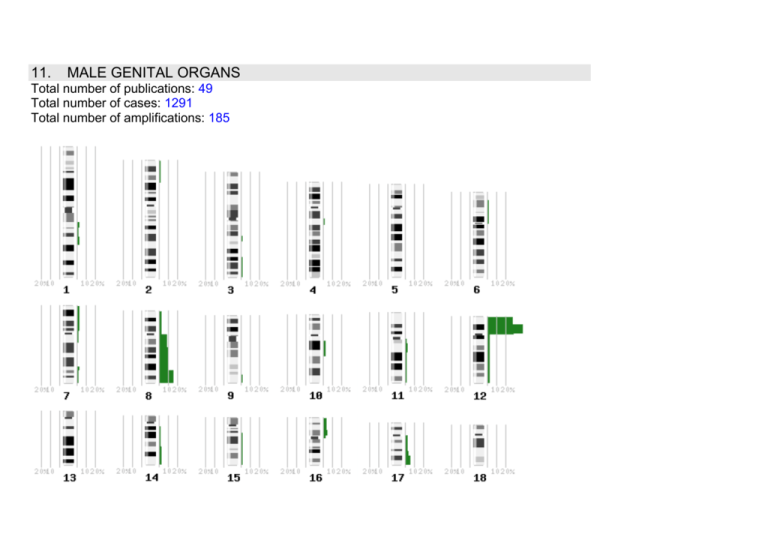
11. MALE GENITAL ORGANS Total number of publications: 49 Total number of cases: 1291 Total number of amplifications: 185 Tumor 11.1 Loss Testis Amplicon 1p32-p36 1p34-pter 2p21-pter 4q12-q21 4q13-qter 5p 5q11.2-q13 5q14-q23 6p11-p22 6q25-qter 7p12-pter 7q 8 9p 9q13-q22 9q31-qter 10 10p12-p13 11p 11q12-q14 Percentage (number of cases) 40 (8/20) 9 (1/11) 9 (1/11) 9 (1/11) 41 (26/63) 43 (15/35) 43 (15/35) 58 (15/26) 9 (1/11) 36 (4/11) 18 (2/11) 9 (1/11) 27 (3/11) 16 (5/31) 25 (5/20) 25 (5/20) 9 (1/11) 25 (5/20) 17 (16/35) 40 (8/20) Amplified genes (studied from the same cases) Reference 1 2 2 2 1-4 1,4 1,4 2,4 2 2 2 2 2 1,2 1 1 2 1 1,4 1 11q22-qter* 12q24.1 13q12-q22 13q31-qter* 16p11.2-p12 16q 17p 18p 18q11.2 18q22-qter 19p 19q 20p 20q11.2-q13.2 22q 63 (26/47) 12p 100 (22/22) 12p11.2-12.1 7 (1/15) 12p11.2-p12.1 91 (10/11) 12p11.23 100 (1/1) 15 (3/20) 46 (16/35) 51 (22/43) 14q12-qter 18 (2/11) 15q15-qter 18 (2/11) 40 (8/20) 16p12-pter 18 (2/11) 15 (3/20) 20 (4/20) 17q24-qter 18 (2/11) 32 (10/31) 40 (8/20) 54 (19/35) 32 (10/31) 19p13.2-pter 36 (4/11) 40 (8/20) 19q13.1-qter 9 (1/11) 15 (3/20) 20 (4/20) 21q11.2-qter 18 (2/11) 35 (7/20) 22q11.2-qter 9 (1/11) Xp11.2-pter 18 (2/11) Xq 9 (1/11) Ypter-q11.2 18 (2/11) 1,2,4,5 1 4 2 6 1 1,4 2-4 2 2 1 2 1 1 2 1,2 1 1,4 1,2 2 1 2 1 1 2 1 2 2 2 2 11.1 TESTICULAR TUMOR (4/61 cases) amp(12p11p12)/ amp(12p11p12)/amp(12p11p12)/amp(12p11p12) Bourdon V, Naef F, Rao PH, Reuter V, Mok SC, Bosl GJ, Koul S, Murty VV, Kucherlapati RS, Chaganti RS: Genomic and expression analysis of the 12p11-p12 amplicon using EST arrays identifies two novel amplified and overexpressed genes. Cancer Res 2002, 62:6218-6223. 11.1 Testis: 76 amplifications out of 65 cases 11.2 Prostate cancer 1p31 1p36.1-pter 2cen-q33 4q13-q31.1 5cen-q31.1 5q14-q23* 6q12-q23* 6q16 8p* 9p21-pter* 10p 10q22-qter* 13q* 15q 16p 16q* 17p* 17q11.2-q24* 18q12-qter* 19 20q 22q Xp11-q13 Xq23-qter 11.2 Prostate cancer Yp Yq11.2 1p34-pter 8q24 (8q) 9q33-qter 12q24 15q15-q21 27 (10/37) 46 (17/37) 23 (18/79) 26 (8/31) 39 (12/31) 21 (19/89) 27 (37/136) 27 (10/37) 60 (81/136) 18 (7/40) 22 (8/37) 29 (34/117) 45 (61/136) 23 (27/117) 14 (7/49) 35 (47/136) 28 (25/89) 12 (7/58) 21 (18/87) 43 (16/37) 15 (13/86) 29 (22/77) 11 (1/9) 11 (1/9) 33 (6/18) 33 (6/18) 50 (8/16) 14 (5/37) 19 (3/16) 25 (4/16) 19 (3/16) 7 7 8-10 8 8-10 8,9 8-11 7 7-11 10 7 7,8 7-11 7-10 9,10 7,8,11 7-10 9 7,10,11 7 9,10 7,10 10 10 9 9 13 7 13 13 13 16 17 19q 20q 22 25 (4/16) 31 (5/16) 38 (6/16) 25 (4/16) 25 (4/16) None Additional reference 11.2 11.2 11.2 13 13 13 13 13 9,12-14 12 PROSTATE CANCER Comment: Expression of androgen receptor gene. High-level amplifications of the AR gene were found in two xenografts (LuCaP 35 and 69), two copies of the chromosome X centromere as well as AR, and in four other xenografts (LuCaP 23.1, 23.8, 23.12, and 70). Linja MJ, Savinainen KJ, Saramäki OR, Tammela TLJ, Vessella RL, Visakorpi T: Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res 2001, 61:35503555. PROSTATE CANCER Comment: Expression of the ERBB2 gene. CISH revealed only one case containing borderline (six to eight copies) amplifications of ERBB2. Savinainen KJ, Saramäki OR, Linja MJ, Bratt O, Tammela TLJ, Isola JJ, Visakorpi T: Expression and gene copy number analysis of ERBB2 oncogene in prostate cancer. Am J Pathol 2002, 160:339-345. PROSTATE CANCER (?/56 cases: 23 primary localized tumors, 18 regional lymph node metastases, and 15 distant metastases) Comment: Number of cases was not clearly defined. High-level amplifications were only seen in metastatic tumors at 1q21, 1q25, 3q21, 4q21, 4q28.3, 7q32, 10q21.3, and Xq12q13. Alers JC, Rochat J, Krijtenburg P-J, Hop WCJ, Kranse R, Rosenberg C, Tanke HJ, Schroder FH, van Dekken H: Identification of genetic markers for prostatic cancer progression. Lab Invest 2000, 80:931-942. 11.2 11.2 11.2 11.2 11.2 PROSTATE CANCER (1/56 cases) amp(8q24) Comment: 0/28 pathologically organ-confined tumors 1/28 tumors with infiltration of the seminal vesicles: amp(8q24) Fu W, Bubendorf L, Willi N, Moch H, Mihatsch MJ, Sauter G, Gasser TC: Genetic changes in clinically organconfined prostate cancer by comparative genomic hybridization. Urology 2000, 56:880-885. PROSTATE CANCER (?/13 cases) Comment: Amplifications were not determined. Verhagen PCMS, Zhu XL, Rohr LR, Cannon-Albright LA, Tavtigian SV, Skolnick MH, Brothman AR: Microdissection, DOP-PCR, and comparative genomic hybridization of paraffin-embedded familial prostate cancers. Cancer Genet Cytogenet 2000, 122:43-48. PROSTATE CANCER (0/10 cases) Sattler H-P, Lensch R, Rohde V, Zimmer E, Meese E, Bonkhoff H, Retz M, Zwergel T, Bex A, Stoeckle M, Wullich B: Novel amplification unit at chromosome 3q25-q27 in human prostate cancer. Prostate 2000, 45:207-215. PROSTATE CANCER (?/2 cases) Comment: Amplifications were not determined. Haapala K, Rökman A, Palmberg C, Hyytinen ER, Laurila M, Tammela TLJ, Koivisto PA: Chromosomal changes in locally recurrent, hormone-refractory prostate carcinomas by karyotyping and comparative genomic hybridization. Cancer Genet Cytogenet 2001, 131:74-78. PROSTATE CANCER (?/60 cases: 40 prostate carcinomas without follow-up and 20 prostate carcinomas with follow-up) Comment: Amplifications were not determined. Mattfeldt T, Wolter H, Kemmerling R, Gottfried H-W, Kestler HA: Cluster analysis of comparative genomic hybridization (CGH) data using self-organizing maps: application to prostate carcinomas. Anal Cell Pathol 2001, 23:29-37. 11.2 11.2 11.2 11.2 11.2 11.2 PROSTATE CANCER (0/21 cases) Rökman A, Koivisto PA, Matikainen MP, Kuukasjärvi T, Poutiainen M, Helin HJ, Karhu R, Kallioniemi O-P, Schleutker J: Genetic changes in familial prostate cancer by comparative genomic hybridization. Prostate 2001, 46:233-239. PROSTATE CANCER (?/27 cases) Comment: Number of cases was not clearly defined, but high-level amplifications were observed at 1q21q25, 1q21q31, 8q21, 8q24, 10q21q22, 10q22, 11q13, 17q23q24, 17q24qter, Xq12q13, Xq13, and Xq13q21. El Gedaily A, Bubendorf L, Willi N, Fu W, Richter J, Moch H, Mihatsch MJ, Sauter G, Gasser TC: Discovery of new DNA amplification loci in prostate cancer by comparative genomic hybridization. Prostate 2001, 46:184190. PROSTATE CANCER (2/50 cases) Wolter H, Gottfried HW, Mattfeldt T: Genetic changes in stage pT2N0 prostate cancer studied by comparative genomic hybridization. BJU Int 2002, 89:310-316. amp(8q24.1qter)/amp(8q24.1qter) PROSTATE CANCER (8/9 cases) amp(16p,17q)/amp(5p,8q,12q,17q)/amp(3q,8q,9q,12q)/amp(16p,17q)/amp(4p,7p,8p)/amp(6q,13q)/ amp(6p,8q,16p)/amp(11q,17q)/amp(11q,12q) Kasahara K, Taguchi T, Yamasaki I, Kamada M, Yuri K, Shuin T: Detection of genetic alterations in advanced prostate cancer by comparative genomic hybridization. Cancer Genet Cytogenet 2002, 137:59-63. PROSTATE CANCER (8/13 cases) amp(2p21pter,7p14q11,8q23qter,16cenp12)/amp(3q26qter,7q32qter)/amp(8cenq21,8q24qter)/amp(8q21q22)/ amp(8q22qter,Xcenq13)/amp(8q23qter)/amp(8q24qter,9q34qter)/amp(8q24qter) Laitinen S, Karhu R, Sawyers CL, Vessella RL, Visakorpi T: Chromosomal aberrations in prostate cancer xenografts detected by comparative genomic hybridization. Genes Chromosomes Cancer 2002, 35:66-73. PROSTATE CANCER (?/18 cases) Comment: Amplifications were not determined. Steiner T, Junker K, Burkhardt F, Braunsdorf A, Janitzky V, Schubert J: Gain in chromosome 8q correlates with early progression in hormonal treated prostate cancer. Eur Urol 2002, 41:167-171. 11.2 11.2 11.2 11.2 11.2 11.2 PROSTATE CARCINOMA (?/20 cases) Comment: Amplifications were not determined. Van Dekken H, Krijtenburg PJ, Alers JC: DNA in situ hybridization (interphase cytogenetics) versus comparative genomic hybridization (CGH) in human cancer: detection of numerical and structural chromosome aberrations. Acta Histochem 2000, 102:85-94. PROSTATE CARCINOMA (?/48 cases) Comment: Amplifications were not determined. Verdorfer I, Hobisch A, Culig Z, Hittmair A, Bartsch G, Erdel M, Duba HC, Utermann G: Combined study of prostatic carcinoma by classical cytogenetic analysis and comparative genomic hybridization. Int J Oncol 2001, 19:1263-1270. PROSTATE CARCINOMA (?/155 cases) Comment: Amplifications were not determined. Schulz WA, Elo JP, Florl AR, Pennanen S, Santourlidis S, Engers R, Buchardt M, Seifert HH, Visakorpi T: Genome wide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer 2002, 35:58-65. PROSTATE CARCINOMAS (?/48 cases) Comment: Amplifications were not determined. Wolter H, Trijic D, Gottfried HW, Mattfeldt T: Chromosomal chances in incidental prostatic carcinomas detected by comparative genomic hybridization. Eur Urol 2002, 41:328-334. PROSTATE ADENOCARCINOMA (?/52 cases) Comment: Amplifications were not determined. Alers JC, Krijtenburg P-J, Vis AN, Hoedemaeker RF, Wildhagen MF, Hop WCJ, van der Kwast TH, Schröder FH, Tanke HJ, van Dekken H: Molecular cytogenetic analysis of prostatic adenocarcinomas from screening studies: early cancers may contain aggressive genetic features. Am J Pathol 2001, 158:399-406. PROSTATE ADENOCARCINOMA (?/33 cases: 16 prostate carcinomas, 5 regional lymph node metastases and 12 prostatic intraepithelial neoplasias ) Comment: Amplifications were not determined. Zitzelsberger H, Engert D, Walch A, Kulka U, Aubele M, Hofler H, Bauchinger M, Werner M: Chromosomal changes during development and progression of prostate adenocarcinomas. Br J Cancer 2001, 84:202-208. 11.2 11.2 PROSTATE TUMORS (0/21 cases) Beheshti B, Vukovic B, Marrano P, Squire JA, Park PC: Resolution of genotypic heterogeneity in prostate tumors using polymerase chain reaction and comparative genomic hybridization on microdissected carcinoma and prostatic intraepithelial neoplasia foci. Cancer Genet Cytogenet 2002, 137:15-22. PROSTATE CANCER REVIEW 11.2 Nupponen NN, Visakorpi T: Molecular cytogenetics of prostate cancer. Microsc Res Tech 2000, 51:456-463. PROSTATE CANCER ADDITIONAL LITERATURE 11.2 Elo JP, Visakorpi T: Molecular genetics of prostate cancer. Ann Med 2001, 33:130-141. PROSTATE CANCER, METASTATIC ADDITIONAL LITERATURE Brown RS, Edwards J, Bartlett JW, Jones C, Dogan A: Routine acid decalcification of bone marrow samples can preserve DNA for FISH and CGH studies in metastatic prostate cancer. J Histochem Cytochem 2002, 50:113-115. 11.2 Prostate tumors: 81 amplifications out of 844 cases 11.3 Spermatocytic seminoma 7 13 15 19 22 X 11.4 11.4 11.4 11.4 Not determined 25 (1/4) 15 50 (2/4) 50 (2/4) 25 (1/4) 50 (2/4) 25 (1/4) 15 15 15 15 15 Testicular germ cell 13 Not determined 100 (3/3) tumor TESTICULAR GERM CELL TUMOUR (2/7 cases) amp(12p11.2p12.1)/amp(12p11.2p12.1) 1 Summersgill B, Osin P, Lu Y-J, Huddart R, Shipley J: Chromosomal imbalances associated with carcinoma in situ and associated testicular germ cell tumours of adolescents and adults. Br J Cancer 2001, 85:213-219. TESTICULAR GERM CELL TUMOUR (?/33 cases: 15 bilateral and 18 sporadic testicular germ cell tumors) Comment: Number of cases was not clearly defined, but high-level amplifications were detected at 12p13 and 12p in bilateral testicular germ cell tumors and at 12p13, 12p12pter, 12p, 14q24qter, and X in sporadic testicular germ cell tumors. Kraggerud SM, Skotheim RI, Szymanska J, Eknaes M, Fossa SD, Stenwig AE, Peltomaki P, Lothe RA: Genome profiles of familial/bilateral and sporadic testicular germ cell tumors. Genes Chromosomes Cancer 2002, 34:168-174. GERM CELL TUMOR (1/6 cases) amp(12q13q14,17q12q21) Comment: 0/2 teratomas 1/4 yolk sac tumors: amp(12q13q14,17q12q21) Mostert M, Rosenberg C, Stoop H, Schuyer M, Timmer A, Oosterhuis W, Looijenga L: Comparative genomic and in situ hybridization of germ cell tumors of the infantile testis. Lab Invest 2000, 80:1055-1064. 11.4 GERM CELL TUMOR, MEDIASTINAL NONSEMINOMATOS (?/35 cases) Comment: Number of cases was not clearly defined, but high-level amplifications were detected at 3p21pter, 12p, 17q11.2q21, 20q, and 21. Schneider DT, Schuster AE, Fritsch MK, Calaminus G, Gobel U, Harms D, Lauer S, Olson T, Perlman EJ: Genetic analysis of mediastinal nonseminomatous germ cell tumors in children and adolescents. Genes Chromosomes Cancer 2002, 34:115-125. 11.4 11.4 11.4 11.4 GERM CELL TUMOR, GONADAL AND NONGONADAL PEDIATRIC (?/42 cases: 21 gonadal and 21 nongonadal) Comment: Amplifications were not determined. Schneider DT, Schuster AE, Fritsch MK, Hu J, Olson T, Lauer S, Gobel U, Perlman EJ: Multipoint imprinting analysis indicates a common precursor cell for gonadal and nongonadal pediatric germ cell tumors. Cancer Res 2001, 61:7268-7276. Undifferentiated tumors None Not determined 0 (0/7) 16 of uncertain origin TESTICULAR SEMINOMA AND NONSEMINOMA (11/11 cases: 4 seminoma and 7 nonseminoma) amp(12p)/amp(12p)/amp(12p)/amp(12p)/amp(12p)/amp(12p)/amp(12p)/amp(12p)/amp(12p)/amp(12p)/amp(1 2p) Comment: Only chromosome 12p sequences were studied. Rosenberg C, Van Gurp RJHLM, Geelen E, Oosterhuis JW, Looijenga LHJ: Overrepresentation of the short arm of chromosome 12 is related to invasive growth of human testicular seminomas and nonseminomas. Oncogene 2000, 19:5858-5862. TESTICULAR SEMINOMA AND NON-SEMINOMA (?/2 cases) Comment: Amplifications were not determined. Looijenga LH, Rosenberg C, van Gurp RJ, Geelen E, van Echten-Arends J, de Jong B, Mostert M, Wolter Oosterhuis J: Comparative genomic hybridization of microdissected samples from different stages in the development of a seminoma and a non-seminoma. J Pathol 2000, 191:187-192. 11.4 Testicular germ cell tumors: 27 amplifications out of 146 cases 11.5 PENILE SQUAMOUS CELL CARCINOMA (1/26 cases) amp(12q13q14) Alves G, Heller A, Fiedler W, Campos MM, Claussen U, Ornellas AA, Liehr T: Genetic imbalances in 26 cases of penile squamous cell carcinoma. Genes Chromosomes Cancer 2001, 31:48-53. Concerning Losses: 10% of the cases must be aberrant and the number of aberrant cases at least three; findings in parentheses are examples of highly frequent aberrations that fail to meet the 3 cases/10% criteria. Boldface indicates 30% aberrant cases detected in a study with at least 10 cases. *Description of a region, e.g. 6q21-q22, implies that in a variety of cases the loss was located within the area but it did not necessarily affect the whole area in all cases. The described regions may therefore not be considered analogous with minimal overlapping area. Furthermore, in some single cases the loss area may extend outside the region described. As a whole, the description should be considered a flexible way to summarize critical areas of recurrent DNA copy number changes in that particular tumor type. Description without an asterisk indicates minimal overlapping areas. Concerning Amplicons: Boldface indicates recurrent established amplicons (at least three cases and frequency more than 5%). REFERENCES 1. Summersgill B, Goker H, Weber-Hall S, Huddart R, Horwich A, Shipley J: Molecular cytogenetic analysis of adult testicular germ cell tumors and identification of regions of consensus copy number change. Br J Cancer 1998, 77:305-313. 2. Korn WM, Olde Weghuis DEM, Suijkerbuijk RF, Schmidt U, Otto T, du Manoir S, Geurts van Kessel A, Harstrick A, Seeber S, Becher R: Detection of chromosomal DNA gains and losses in testicular germ cell tumors by comparative genomic hybridization. Genes Chromosomes Cancer 1996, 17:78-87. 3. Mostert MMC, van de Pol M, Olde Weghuis D, Suijkerbuijk RF, Geurts van Kessel A, van Echten J, Oosterhuis JW, Looijenga LHJ: Comparative genomic hybridization of germ cell tumors of the adult testis: confirmation of karyotypic findings and identification of a 12p-amplicon. Cancer Genet Cytogenet 1996, 89:146-152. 4. Ottesen AM, Kirchhoff M, De-Meyts ER, Maahr J, Gerdes T, Rose H, Lundsteen C, Petersen PM, Philip J, Skakkebaek NE: Detection of chromosomal aberrations in seminomatous germ cell tumors using cmparative genomic hybridization. Genes Chromosomes Cancer 1997, 20:412-418. 5. Speicher MR, Jauch A, Walt H, du Manoir S, Ried T, Jochum W, Sulser T, Cremer T: Correlation of microscopic phenotype with genotype in a formalin-fixed, paraffin-embedded testicular germ cell tumor with universal DNA amplification, comparative genomic hybridization, and interphase cytogenetics. Am J Pathol 1995, 146:1332-1340. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. Suijkerbuijk RF, Sinke RJ, Olde Weghuis DEM, Roque L, Forus A, Stellink F, Siepman A, van de Kaa C, Soares J, Geurts van Kessel A: Amplification of chromosome subregion 12p11.2-p12.1 in a metastasis of an i(12p)-negative seminoma: relationship to tumor progression? Cancer Genet Cytogenet 1994, 78:145-152. Nupponen NN, Kakkola L, Koivisto P, Visakorpi T: Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol 1998, 153:141-148. Cher ML, Bova GS, Moore DH, Small EJ, Carroll PR, Pin SS, Epstein JI, Isaacs WB, Jensen RH: Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res 1996, 56:3091-3102. Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen RH: Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes Chromosomes Cancer 1994, 11:153-162. Visakorpi T, Kallioniemi AH, Syvänen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi O-P: Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995, 55:342-347. Joos S, Bergerheim USR, Pan Y, Matsuyama H, Bentz M, du Manoir S, Lichter P: Mapping of chromosomal gains and losses in prostate cancer by comparative genomic hybridization. Genes Chromosomes Cancer 1995, 14:267-276. Cher ML, Lewis PE, Banerjee M, Hurley PM, Sakr W, Grignon DJ, Powell IJ: A similar pattern of chromosomal alterations in prostate cancers from African-Americans and Caucasian Americans. Clin Cancer Res 1998, 4:1273-1278. Sattler HP, Rohde V, Bonkhoff H, Zwergel T, Wullich B: Comparative genomic hybridization reveals DNA copy number gains to frequently occur in human prostate cancer. Prostate 1999, 39:79-86. Koivisto PA, Schleutker J, Helin H, Ehren-van Eekelen C, Kallioniemi O-P, Trapman J: Androgen receptor gene alterations and chromosomal gains and losses in prostate carcinomas appearing during finasteride treatment for benign prostatic hyperplasia. Clin Cancer Res 1999, 5:3578-3582. Rosenberg C, Mostert MC, Schut TB, van de Pol M, van Echten J, de Jong B, Raap AK, Tanke H, Oosterhuis JW, Looijenga LHJ: Chromosomal constitution of human spermatocytic seminomas: comparative genomic hybridization supported by conventional and interphase cytogenetics. Genes Chromosomes Cancer 1998, 23:286-291. Summersgill B, Goker H, Osin P, Huddart R, Horwich A, Fisher C, Shipley J: Establishing germ cell origin of undifferentiated tumors by identifying gain of 12p material using comparative genomic hybridization analysis of paraffin-embedded samples. Diagn Mol Pathol 1998, 7:260-266.