Protein Sequencing with a Modifed Edman Reagent
advertisement

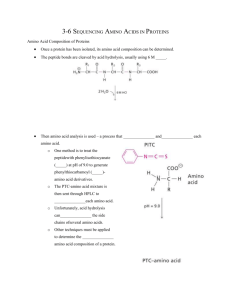
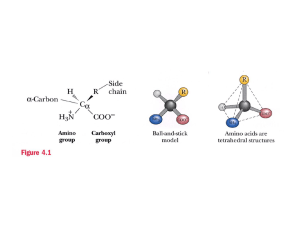
Topics in Nanobiotechnology 2004 MV Duarte Edman sequencing Edman sequencing The conventional Edman sequencing- Determination of aminoacid sequence in peptides or protetins: The amino terminus of a protein or polypeptide is first derivatized with phenylisothiocyanate (PITC) under basic conditions (triethylamine (TEA) or diisoproylethylamine (DIEA) ). This coupling step produces a phenylthiocarbamyl (PTC) peptide or protein. The thiocarbonyl function of the PTC is a moderately strong nucleophile, and under acidic conditions it will attack the carbonyl carbon of the adjacent peptide bond. This nucleophilic attack or cleavage step results in the production of an anilothiazolinone (ATZ) of the terminal amino acid and leaves the original peptide or protein shortened by exactly one amino acid residue. The ATZ of terminal amino acid has different solubility properties from the peptide or protein, so it can be extracted and subjected to further analysis. The shortened peptide or protein again has a bare amino terminus, so it can be subjected to additional cycles of coupling, cleavage, and extraction. The extracted ATZ of the terminal amino acid, however, is not stable. Under acidic aqueous conditions, ATZ's rearrange rapidly to form more stable phenylthiohydantoins (PTH's), which are more amenable to analysis. The stable PTH's are then analyzed by UV absorption detection reverse phase High Performance Liquid Chromatography (UV/HPLC). Roger Moore from The Beckman Research Institute is a graduate student working in modifications of the technique. I found his explanation very interesting because He describes the technique, as a technique for determining aminoacid sequences by a serie of reactions where pH control and well selected reactive make possible to cut a protein or peptide amino acid by aminoacid for further characterization. But explain a negative point of the conventional technique, which is the final method for characterization of the aminoacid (UV/HPLC) , because this methods can not get to the lower concentration that biologists need nowadays. He explained that two ways of improving the method were studied, Fluorescence detection (but did not work because of kinetics problems when changing PITC of other fluorescent isothiocyanates) and Mass Spectrometry (wich did not work neither because when putting some reactive readily protonable or deprotonatable or with a fixed positive or negative charge center, problems of solubility appeared resulting in ATZ solubility similar to protein so it was impossible to separate) He and his group are working to improved the technique by sensitivity and detection speed using MS but developing a compound witch can be detected by MS but its solubility from the shortened protein is different enough to separate and characterize it. Unfortunately, at the moment, I did not found any new results of this man´s work. ____________________________________________________________________ Protein Sequencing with a Modifed Edman Reagent- by Roger Moore Introduction My project at work is basically an attempt to develop and improved method of chemical protein sequencing by modifying the current state of the art technique. This is considerably more difficult than it sounds. The current state of the art is the result of Topics in Nanobiotechnology 2004 MV Duarte Edman sequencing literally decades of work improving a single basic methodology, the Edman Method. Dozens of alternative techniques for chemical protein sequencing have been tried, but none have caught on. The Edman Technique In order to understand why my work is necessary, it's necessary to look at the advantages and limitations of the current, Edman, methodology. In Edman sequencing, the amino terminus of a protein or polypeptide is first derivatized with phenylisothiocyanate (PITC) under basic conditions. Because PITC is a good electrophile, the base must be non-nucleophilic, so a tertiary amine such as triethylamine (TEA) or diisoproylethylamine (DIEA) is used. This coupling step produces a phenylthiocarbamyl (PTC) peptide or protein. The thiocarbonyl function of the PTC is a moderately strong nucleophile, and under acidic conditions it will attack the carbonyl carbon of the adjacent peptide bond. This nucleophilic attack or cleavage step results in the production of an anilothiazolinone (ATZ) of the terminal amino acid and leaves the original peptide or protein shortened by exactly one amino acid residue. The ATZ of terminal amino acid has different solubility properties from the peptide or protein, so it can be extracted and subjected to further analysis. The shortened peptide or protein again has a bare amino terminus, so it can be subjected to additional cycles of coupling, cleavage, and extraction. The extracted ATZ of the terminal amino acid, however, is not stable. Under acidic aqueous conditions, ATZ's rearrange rapidly to form more stable phenylthiohydantoins (PTH's), which are more amenable to analysis. The stable PTH's are then analyzed by UV absorbtion detection reverse phase High Performance Liquid Chromatography (UV/HPLC). The Problem The Edman technique has a number of great strengths. From a chemical standpoint, the greatest strenght is the ability to separate the reaction steps by changing the acid/base conditions. This allows very careful control over the reaction so that the protein or peptide is degraded exactly one amino acid at a time. From a more practical standpoint, the greatest strength is the tremendous number of man years of work which have gone into perfecting each and every step of the reaction. Different reagents have been compared for their efficiency in each step of the reaction, and other reaction conditions have been heavily optimized. Very careful choice of the conditions and reagents has lead to a very well developed methodology. In some ways, though, the heavy investment in the methodology can be seen as a disadvantage. Each and every aspect of Edman sequencing, from the preparation of the reagents through the detector at the end of the chromatograph has been tweaked to give the best possible results. Unfortunately, this means that it's highly unlikely that the method can get much better without a fundamental change in either its technology or the chemistry. Advances in biological technique have pushed beyond the ability of current Edman sequencing to handle, and small improvements won't do. The weak point in Edman sequencing is the UV/HPLC detector. UV absorbtion detectionjust isn't able to detect PTH amino acids below the picomole level, and biologists are now able to isolate and purify femtomoles of proteins and peptides. This means that the most interesting targets for sequencing are available in quanities too small to sequence chemically. Others' Attempts This problem has not, of course, gone unnoticed. Even before detection sensitivity problems made improvement to the Edman technique necessary numerous attempts were made to improve on the basic Edman scheme. To date, none of them has succeeded. There are two obvious ways of improving the detection limits of Edman sequencing, and both have been tried extensively. The first method is fluorescence detection. Fluorescence is potentially much more sensitive than UV absorbtion as a HPLC detecition technique, and many fluorescence detectors are commercially available. Furthermore, there are already a number of well characterized fluorescent isothiocyanates which can be easily used in place of PITC in the reaction scheme. Unfortunately, these attempts have run into difficulties with reaction kinetics. The isothiocyanate used in Edman-type sequencing reacts first as an electrophile and then as a nucleophile, so any electronic effects from its side chain work in opposite directions in the coupling and cleavage steps. PITC has turned out to be the ideal choice for sequencing because it is roughly as reactive in coupling as it is in cleavage. Proposed alternates (with the exception of methyl isothiocyanate and related aliphatic isothiocyanates, which have had some success) have generally accelerated one of the two reactions greatly, but slowed the other by an equal amount. The slower reaction, though, is enough slower so that the overall reaction is too slow to be of practical use. The other obvious detection technique is mass spectrometry (MS). Like fluorescence, MS is much more sensitive than UV absorbtion, and it adds the advantage of being very much faster than HPLC. Modern developments in ionization, such as electrospray, fast atom bombardment, secondary ion mass spectrometry, and matrix assisted laser desorbtion ionization, have made MS of biomolecules a comparatively straightforward matter. The problems encountered with MS have been somewhat more complicated than those in fluorescence. For modern MS ionization methods to work, a molecule must have a readily protonatable or deprotonatable group, or have a fixed positive or negative charge center, such as a quaternary amine. These groups, however, are generally quite polar, so that cleaved derivatives (analagous to ATZ's) containing them wind up having solubility properties close enough to those of proteins and peptides that extracting them is impracticable. To avoid this problem, later attempts often tried to incorporate non-polar functional groups which could be derivatized to give ionizable functions after extraction. These reactive groups have had tended to react to give a variety of intractable side products while still reacting with the proteins or peptides, which has prevented them from being usable. Our First Plan Topics in Nanobiotechnology 2004 MV Duarte Edman sequencing My work (with Drs. Stan Hefta, Terry Lee, and John Shively in the Department of Immunology, Beckman Research Institute, City of Hope) has focused on MS detection as a method of improving sensitivity and detection speed simultaneously. It's hoped that the improvement in detection speed will allow a single MS detector to work with several samples in paralell, speeding overall sequencing throughput while still giving improved sensitivity. Our first attempt used a modified secondary derivitization method. Rather than incorporating a separate functional group which would be derivatized later, we tried using a sequencing reagent other than an isothiocyanate. Instead, we used an activated thiobenzoic acid derivative. This coupled and cleaved essentially the same way that PITC does, but it's PhTZ derivatives (analagous to ATZ's) could not rearrange to give a molecule analagous to a PTH. This left the molecule relatively reactive, and it could be derivatized with a bifunctional amine to give a readily ionizable derivative. While promising, this method ran into unexpected difficulties with the PhTZ's solubility properties. They turned out to be very similar in solubility to peptides, so that it wasn't possible to separate them. After considerable work trying to resolve this problem, we were forced to abandon this methodology. My Current Work I'm now working on a new reagent which incorporates a tertiary amine as a protonatable site. By using a tertiary amine with comparatively long aliphatic side chains, we've discovered that it's possible to get enough differential solubility to make exracting our ATZ analog work. The final derivatives have shown excellent mass spectral sensitivity, which make us optomistic about out long term prospects. Synthesizing my new reagent has been difficult. Isothiocyanates are generally annoying synthetic targets, as all of the available methods of synthesizing them involve either unpleasant reagents or nasty biproducts, or both. My reagent has been particularly difficult. The bifunctional amine I used in the synthesis had a tendency to hydrogen bond to itself, which resulted in its being a much more reacive nucleophile than expected. That meant that it tended to form undesirable thioureas instead of the desired isothiocyanates. I was eventually able to solve this problem by adding one equivalent of acid to the amine before starting the reaction. The acid prevented the amine from hydrogen bonding to itself, reducing its reactivity and making the reaction run as planned. Purification by vacum distillation was tedious, but not particularly difficult. Now that I have succeeded in synthesizing and purifying my new reagent, I'm attempting to work out acceptable reaction conditions for the use in the sequencer. I expect this to take several more months. http://alumnus.caltech.edu/~raj/work.html