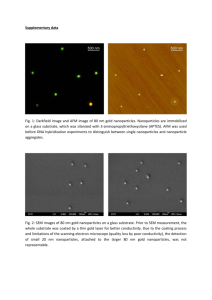
SUPPLEMENTARY MATERIAL
advertisement

SUPPLEMENTARY INFORMATION Effect of PEG biofunctional spacers and TAT peptide on dsRNA loading on gold nanoparticles Vanesa Sanz1, João Conde2,1, Yulán Hernández1, Pedro V. Baptista2, M.R. Ibarra1 and Jesús M. de la Fuente1* 1 Instituto de Nanociencia de Aragón, Universidad de Zaragoza, Mariano Esquillor s/n 50018, Zaragoza, Spain. 2 Centro de Investigação em Genética Molecular Humana (CIGMH), Departamento de Ciências da Vida, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, 2829-516 Caparica, Portugal. Figure S1 – Citrate-Gold nanoparticle characterization. TEM images (left, scale bar = 100 nm), size distribution histogram (right) showing an average diameter of 14.47 nm. 1 Figure S2 – A. TEM image of AuNPs@COOH (scale bar: 100 nm). B. TEM image of AuNPs@COOH/N3 (scale bar: 100 nm). C. UV/Vis Spectroscopy of AuNPs@COOH/N3, AuNPs@COOH/N3@TAT and AuNPs@COOH/N3@TAT@dsRNA. 2 1. Determination of the degree of saturation of gold nanoparticles functionalized with thiolated polyethyleneglycol chains. For the determination of the degree of saturation of gold nanoparticles functionalized with thiolated polyethyleneglycol chains we developed and optimized a procedure based on the Ellman’s assay. With this procedure it is possible to determine the number of thiolated chains that have been bound to gold nanoparticles. After the incubation of the gold nanoparticles with the thiolated chains, the nanoparticles are functionalized with a defined number of chains being the excess of thiolated chains free in solution. In order to determine the number of bound chains, this suspension is centrifuged and the supernatant with the excess of thiolated chains is kept to perform the quantification assay. The number of exchanged chains is given by the difference between the amount determined by this assay and the initial amount of chains in the incubation mixture with the nanoparticles. As it was stated above, the free thiolated chains are determined by a procedure based on the Ellman’s assay. In this assay, the supernatant, containing the spacer with thiol groups on its structure, reacts with DTNB (5,5’-dithio-bis(2-nitrobenzoic) acid) to give a colored product that can be measured spectrophotometrically. In the optimal conditions, 200 μL of stock solution of the thiolated chain or the sample containing the supernatant are mixed with 100 μL of phosphate buffer 0.5 M pH 7 and 7 μL of DTNB 5 mg/mL in phosphate buffer 0.5 M pH 7. The mixture is made to react for 10 minutes and the absorbance at 412 nm is measured. The linear range for the thiolated chain is 0.5-30 μg/mL (Abs412 = 0.0229[chain, µg/mL] + 0.0587). The total number of thiolated chains that can be attached per gold nanoparticle were calculated. As it is shown in Figure S3, the excess of thiolated chain versus the initial concentration of thiolated chain in an incubation mixture with a constant concentration of gold nanoparticles is represented. As the initial concentration of thiolated chain increases, the excess of free chain increases in an equilibrium dependent way (equilibrium constant [bound chain]/[free chain]=0.76). There is a point at which the 3 nanoparticle becomes saturated with a thiolated layer and is not able to uptake more thiolated chains. At this point it is possible to calculate the maximum number of thiolated chains that it is possible to bind per gold nanoparticle being estimated a number of around 5000 chains per nanoparticle. Figure S3. Variation of the excess of thiolated chain as a function of the initial concentration in the incubation mixture with gold nanoparticles. Initial concentration of gold nanoparticles 0.5 mg/mL. Therefore, it is possible to prepare gold nanoparticles with a controlled degree of saturation if an initial thiolated chain concentration below the saturation conditions is used (below 0.05 mg/mL of thiolated chain per 0.5 mg/mL of gold nanoparticles). In order to determine the degree of saturation at which gold nanoparticles are stable enough and allow the binding of additional thiolated chains we performed the following assay. Gold nanoparticles were functionalized with a controlled amount of thiolated chains in order to obtain nanoparticles with different degrees of saturation. After this, a fixed amount of a thiolated chain was added and incubated with the gold nanoparticles. The 4 concentration of thiolated chain that was bound to the non-saturated gold nanoparticles was determined by the Ellman’s assay. As it is shown in Figure S4, as the initial percentage of saturation of the thiolated layer decreases the gold nanoparticles are able to bind a higher amount of thiolated chain added on this second addition. The best conditions for the binding by a second addition of a thiolated molecule are to work with gold nanoparticles with a degree of saturation below 35 %. However, gold nanoparticles with a degree of saturation below 10 % are less stable. For these reasons, the best conditions for study the binding of thiolated molecules on gold nanoparticles functionalized with a non-saturated thiolated chain layer is between 10 and 35 %. Figure S4. Determination of the amount of thiolated chain bound to gold nanoparticles already functionalized with a thiolated chain layer with different degree of saturation. The initial concentration of gold nanoparticles is 0.5 mg/mL. 5 2. Procedure for the determination of the bound TAT peptide by the EDC coupling reaction to functionalized gold nanoparticles. As it was stated in method section, a procedure for the determination of the peptide that was bound to gold nanoparticles through the EDC coupling reaction was developed. This assay was based on the Bradford method. Given that the reagents for the EDC reaction (sulfo-NHS, EDC) interferes with the assay an standard addition calibration curve for the TAT peptide in the EDC reaction conditions was obtained (see Figure S5). This calibration curve was used for the interpolation of the samples containing the excess of peptide that was not bound to gold nanoparticles. Figure S5. Standard addition calibration curve for the TAT peptide in the conditions of the EDC coupling reaction. In the linear range the calibration curve is given by the following equation Abs570 = 0.071[peptide, μg/mL] + 0.24. 6 3. Quantification of the dsRNA strand loaded into gold nanoparticles In order to carry out the dsRNA quantification we used the GelRed™ dye as a nucleic acid intercalator. The GelRed™ is a sensitive, stable fluorescent nucleic acid dye commonly designed to stain dsDNA, ssDNA as well as ds and ssRNA. Here, we had developed an analytical method to quantify the amount of dsRNA that was lost during the functionalization. We measured the dsRNA concentration in the supernatants after incubation, which is measuring the dsRNA in excess, that hadn’t bound to gold surface. Briefly, the supernatants were incubated with GelRed 100× and TBE 0.5× and fluorescence was measured with emission at 602 nm. A Correlation to a standard curve of known dsRNA concentrations allows accurate measurement of dsRNA in unknown samples. In Figure S6A we can see the fluorescence spectra of the GelRed only and when it is bound to dsRNA. Figure S6B presents the calibration curve for the thiolated dsRNA used in the experiments. Figure S6. A. Fluorescence spectra of the GelRed only and dsRNA-GelRed. B. Calibration curve for the thiolated dsRNA. In the linear range the calibration curve is given by the following equation: Emission at 602nm = 439.96[dsRNA, nmol] + 32.697. 7