Supplemental Table 4: Locus information, gene names used
advertisement
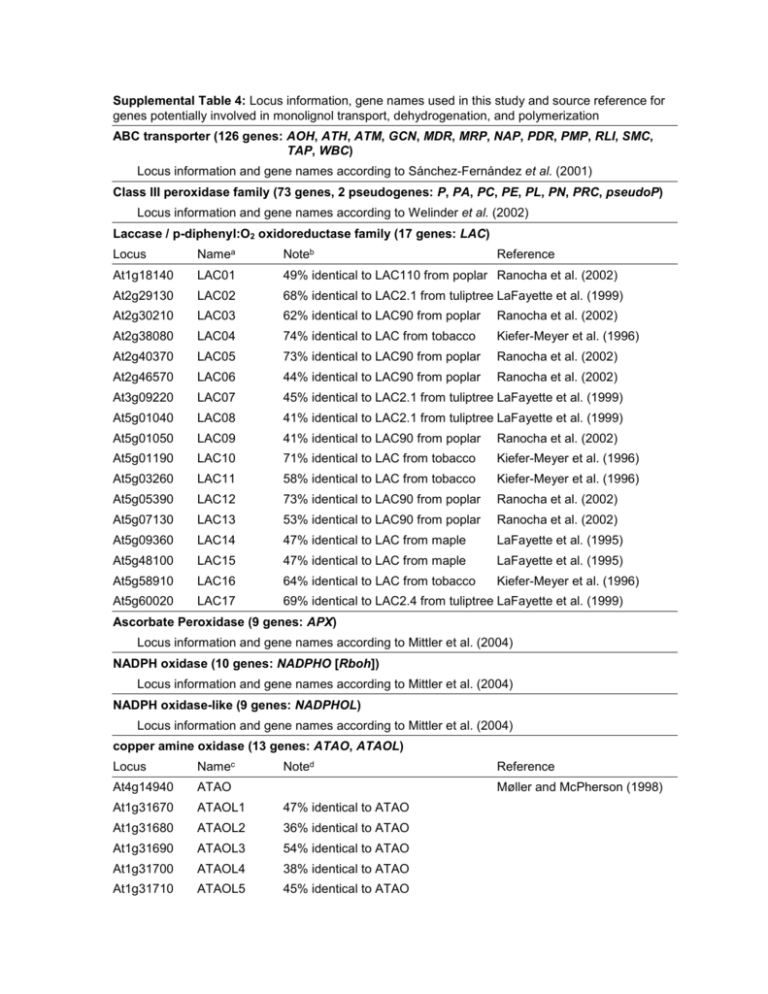
Supplemental Table 4: Locus information, gene names used in this study and source reference for genes potentially involved in monolignol transport, dehydrogenation, and polymerization ABC transporter (126 genes: AOH, ATH, ATM, GCN, MDR, MRP, NAP, PDR, PMP, RLI, SMC, TAP, WBC) Locus information and gene names according to Sánchez-Fernández et al. (2001) Class III peroxidase family (73 genes, 2 pseudogenes: P, PA, PC, PE, PL, PN, PRC, pseudoP) Locus information and gene names according to Welinder et al. (2002) Laccase / p-diphenyl:O2 oxidoreductase family (17 genes: LAC) Locus Namea Noteb At1g18140 LAC01 49% identical to LAC110 from poplar Ranocha et al. (2002) At2g29130 LAC02 68% identical to LAC2.1 from tuliptree LaFayette et al. (1999) At2g30210 LAC03 62% identical to LAC90 from poplar Ranocha et al. (2002) At2g38080 LAC04 74% identical to LAC from tobacco Kiefer-Meyer et al. (1996) At2g40370 LAC05 73% identical to LAC90 from poplar Ranocha et al. (2002) At2g46570 LAC06 44% identical to LAC90 from poplar Ranocha et al. (2002) At3g09220 LAC07 45% identical to LAC2.1 from tuliptree LaFayette et al. (1999) At5g01040 LAC08 41% identical to LAC2.1 from tuliptree LaFayette et al. (1999) At5g01050 LAC09 41% identical to LAC90 from poplar Ranocha et al. (2002) At5g01190 LAC10 71% identical to LAC from tobacco Kiefer-Meyer et al. (1996) At5g03260 LAC11 58% identical to LAC from tobacco Kiefer-Meyer et al. (1996) At5g05390 LAC12 73% identical to LAC90 from poplar Ranocha et al. (2002) At5g07130 LAC13 53% identical to LAC90 from poplar Ranocha et al. (2002) At5g09360 LAC14 47% identical to LAC from maple LaFayette et al. (1995) At5g48100 LAC15 47% identical to LAC from maple LaFayette et al. (1995) At5g58910 LAC16 64% identical to LAC from tobacco Kiefer-Meyer et al. (1996) At5g60020 LAC17 69% identical to LAC2.4 from tuliptree LaFayette et al. (1999) Reference Ascorbate Peroxidase (9 genes: APX) Locus information and gene names according to Mittler et al. (2004) NADPH oxidase (10 genes: NADPHO [Rboh]) Locus information and gene names according to Mittler et al. (2004) NADPH oxidase-like (9 genes: NADPHOL) Locus information and gene names according to Mittler et al. (2004) copper amine oxidase (13 genes: ATAO, ATAOL) Locus Namec Noted At4g14940 ATAO At1g31670 ATAOL1 47% identical to ATAO At1g31680 ATAOL2 36% identical to ATAO At1g31690 ATAOL3 54% identical to ATAO At1g31700 ATAOL4 38% identical to ATAO At1g31710 ATAOL5 45% identical to ATAO Reference Møller and McPherson (1998) At1g62810 ATAOL6 41% identical to ATAO At2g42490 ATAOL7 23% identical to ATAO At3g43670 ATAOL8 39% identical to ATAO At3g43820 ATAOL9 35% identical to ATAO At4g12270 ATAOL10 28% identical to ATAO At4g12280 ATAOL11 45% identical to ATAO At4g12290 ATAOL12 42% identical to ATAO Oxalate oxidase, germin like proteins (32 genes: GLP) Locus Namee Noteb At1g72610 GLP1 Carter et al. (1998) At5g39160 GLP2a Carter et al. (1998) At5g39190 GLP2c 99% identical to GLP2a At5g39130 GLP2d 96% identical to GLP2a At5g20630 GLP3 Carter et al. (1998) At1g18970 GLP4 Carter et al. (1998) At1g18980 GLP4b 84% identical to GLP4 At1g74820 GLP4c 55% identical to GLP4 At1g09560 GLP5 Carter et al. (1998) At5g39100 GLP6a Carter et al. (1998) At5g39110 GLP6b 95% identical to GLP6 At5g39120 GLP6c 95% identical to GLP6 At5g39150 GLP6e 95% identical to GLP6 At5g39180 GLP6f 94% identical to GLP6 At1g10460 GLP7 Carter et al. (1998) At3g05930 GLP8 Carter et al. (1998) At5g26700 GLP8b At4g14630 GLP9a At5g38940 GLP9b 72% identical to GLP9 At5g38930 GLP9c 72% identical to GLP9 At5g38910 GLP9e At5g38960 GLP9d At3g62020 GLP10 At3g04150 GLP11a 55% identical to GLP6 At3g04170 GLP11b 72% identical to GLP11a At3g04180 GLP11c 72% identical to GLP11a At3g04190 GLP11d 72% identical to GLP11a At3g04200 GLP11e 62% identical to GLP11a At3g05950 GLP12 79% identical to GLP6 At3g10080 GLP13 43% identical to GLP4 Reference 64% identical to GLP8 Carter et al. (1998) Carter et al. (1998) 69% identical to GLP9 Carter et al. (1998) At5g38950 GLP14 48% identical to GLP2a At5g61750 GLP15 35% identical to GLP5 Dirigent like proteins (21 genes: DIR) Locus Namef Noteg Reference At5g42510 DIR1 24 % identical to DIR from Forsythia Davin et al. (1997) At5g42500 DIR2 27 % identical to DIR from Forsythia Davin et al. (1997) At5g49040 DIR3 21 % identical to DIR from Forsythia Davin et al. (1997) At2g21110 DIR4 24 % identical to DIR from Forsythia Davin et al. (1997) At1g64160 DIR5 45 % identical to DIR from Forsythia Davin et al. (1997) At4g23690 DIR6 47 % identical to DIR from Forsythia Davin et al. (1997) At3g13650 DIR7 23 % identical to DIR from Forsythia Davin et al. (1997) At3g13662 DIR8 22 % identical to DIR from Forsythia Davin et al. (1997) At2g39430 DIR9 24 % identical to DIR from Forsythia Davin et al. (1997) At2g28670 DIR10 27 % identical to DIR from Forsythia Davin et al. (1997) At1g22900 DIR11 27 % identical to DIR from Forsythia Davin et al. (1997) At4g11180 DIR12 37 % identical to DIR from Forsythia Davin et al. (1997) At4g11190 DIR13 39 % identical to DIR from Forsythia Davin et al. (1997) At4g11210 DIR14/16 39 % identical to DIR from Forsythia Davin et al. (1997) At4g38700 DIR15 25 % identical to DIR from Forsythia Davin et al. (1997) At4g13580 DIR18 23 % identical to DIR from Forsythia Davin et al. (1997) At1g55210 DIR19 22 % identical to DIR from Forsythia Davin et al. (1997) At1g58170 DIR20 24 % identical to DIR from Forsythia Davin et al. (1997) At1g65870 DIR21 27 % identical to DIR from Forsythia Davin et al. (1997) At2g21100 DIR22 29 % identical to DIR from Forsythia Davin et al. (1997) At3g13660 DIR23 27 % identical to DIR from Forsythia Davin et al. (1997) a gene names are designated LAC followed by a number in order of the Locus identifier amino acid identities based on a DiAlign alignment to the closest phylogenetic relative that has been characterized c gene names similar to ATAO (Møller and McPherson, 1998)are designated ATAOL followed by a number in order of the Locus identifier d amino acid identities based on a DiAlign alignment to ATAO (Møller and McPherson, 1998) e gene names follow the system introduced by Carter et al., (1998) f gene names as assigned by Norman G. Lewis (ibc.wsu.edu/research/lewis/nsf/index.html) were applicable (DIR1 to DIR 18), then in order of the Locus identifier b References for Supplemental Table 4 Carter, C., Graham, R.A. and Thornburg, R.W. (1998) Arabidopsis thaliana contains a large family of germin-like proteins:characterization of cDNA and genomic sequences encoding 12 unique family members. Plant Mol. Biol. 38, 929–943. Davin, L.B., Wang, H.-B., Crowell, A.L., Bedgar, D.L., Martin, D.M., Sarkanen, S. and Lewis, N.G. (1997) Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362-367. Kiefer-Meyer, M.-C., Gomord, V., O'Connell, A., Halpin, C. and Faye, L. (1996) Cloning and sequence analysis of laccase-encoding cDNA clones from tobacco. Gene 178, 205-207. LaFayette, P.R., Eriksson, K. and Dean, J. (1995) Nucleotide sequence of a cDNA clone encoding an acidic laccase from Sycamore Maple (Acer pseudoplatanus L.). Plant Physiol. 107, 667-668. LaFayette, P.R., Eriksson, K.-E.L. and Dean, J.F.D. (1999) Characterization and heterologous expression of laccase cDNAs from xylem tissues of yellow-poplar (Liriodendron tulipifera). Plant Mol. Biol. 40, 23–35. Mittler, R., Vanderauwera, S., Gollery, M. and Van Breusegem, F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490-498. Møller, S.G. and McPherson, M.J. (1998) Developmental expression and biochemical analysis of the Arabidopsis atao gene encoding an H2O2 generating diamine oxidase. Plant J. 13, 781-791. Ranocha, P., Chabannes, M., Chamayou, S., Danoun, S., Jauneau, A., Boudet, A.-M. and Goffner, D. (2002) Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in Poplar. Plant Physiol. 129, 145-155. Sánchez-Fernández, R., Davies, T.G.E., Coleman, J.O.D. and Rea, P.A. (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 276, 30231-30244. Welinder, K.G., Justesen, A.F., Kjaersgard, I.V.H., Jensen, R.B., Rasmussen, S.K., Jespersen, H.M. and Duroux, L. (2002) Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 269, 60636081.