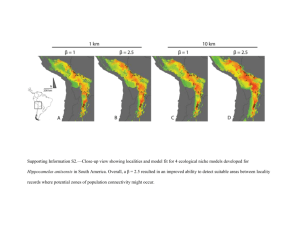
Supplemental data include detailed methods for Functional
advertisement

Supplemental Data
Feature-reduction and semi-simulated data in functional
connectivity-based cortical parcellation
Xiaoguang Tian2,*, Cirong Liu2,5,*, Tianzi Jiang4,5, Joshua Rizak2, Yuanye Ma1,2,3,6, Xintian
Hu1,2,3,6
1
Yunnan Key Lab of primate Biomedical Research, China
2
Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan, China
3
State Key Laboratory of Brain and Cognitive Science, Institute of Biophysics, Chinese
Academy of Sciences, Beijing, China
4
LIAMA Center for Computational Medicine, National Laboratory of Pattern Recognition,
Institute of Automation, Chinese Academy of Sciences, Beijing, China
5
The University of Queensland, Queensland Brain Institute, QLD 4072, Australia
6
Kunming Bio-International
Functional connectivity maps by different feature reduction
approaches
Based on Principal Component Analysis (PCA):
The feature reduction approach by PCA differed from the approach by Affinity
Propagation (AP) as it was directly done on the raw functional connectivity maps. For
example, suppose we let B= n × p represent seed regions’ functional connectivity maps
of n samples (seed regions’ voxels) under p variables (features of each one seed region
voxel), as:
Bn p
x11
x
n ,1
x1, p
xn , p
(1)
The first step required a subtraction of the mean of the variables under each of the sample.
i ,: xi ,: mi
Where, the mean of variables mi
(2)
1 p
xi ,:
p i 1
Thus, the whole adjusted matrix was represented as:
Wn p
11
n ,1
1, p
n , p
(3)
Secondly, the above adjusted matrix was transferred into a covariance matrix Cp p W TW
C p p
cov(1,1)
W W
cov( p,1)
T
cov(1, p)
cov( p, p)
(4)
Then, the eigenvalues and eigenvectors from the covariance matrix were obtained by
singular value decomposition (SVD). However, the number of eigenvectors
corresponding to the non-zero eigenvalues determined by the SVD was the number of
samples n subtracted by 1, because the experimental data for the number of samples
(voxels) n was less than the number of variables p (features). Lastly, if we select r
satisfied eigenvectors, the functional connectivity maps after PCA would be created as:
B 'nr Wn p E pr
where B’ was the final functional connectivity maps and E was eigenvector matrix.
(5)
In this experiment, we averaged 25 subjects’ raw seed region functional connectivity
maps to obtain the group functional connectivity maps and then performed the above
feature reduction by PCA on the group functional connectivity maps. We select all
principal components (PCA-all) and principal components covering 95% cumulative
contribution (PCA-95) to build new functional connectivity maps.
Based on Affinity Propagation:
As illustrated in Fig. 1, the feature reduction based on Affinity Propagation first applied a
gross parcellation of all voxels within the whole brain. We used the standard Affinity
Propagation algorithm
implemented in Matlab and freely available through
http://www.psi.toronto.edu/index.php?q=affinity%20propagation
[1]
.
The
functional
connectivity maps of the whole brain were transformed into a collection of negativevalued similarities in order to satisfy a special inputting format needed for Affinity
Propagation algorithm. Briefly, there were two elements xi , xk . The negative-valued
similarity between them was given by s(i, k ) || xi xk || . This transformation allowed
Affinity Propagation to automatically receive the best number of clusters (K) setting by a
certain preference, rather than by pre-specifying an arbitrary number of clusters.
Nonetheless, this preference needed to be set to a common value. The value was defined
as the median of the input similarities that resulted in a moderate number of clusters as
described by Frey and Dueck
[1]
. Then Affinity Propagation used a process of iteration
under fixed maximal numbers (max=2000), where two kinds of message were exchanged
between data points. The first type of message, termed “Responsibility” r (i, k ) reflected
the accumulated evidence for how well-suited a point k was to serve as the exemplar for
point i , while taking into account all other potential exemplars for point i . The message
was updated following the rule:
r (i, k ) s(i, k ) max{a(i, k ') s(i, k ')}
k ' k
(6)
Another message, termed “Availability” a(i, k ) , was sent from candidate exemplar point
k to point i , and reflected the accumulated evidence for how appropriate it would be for
point i to choose point k as its exemplar, taking into account the support from all other
points that point k should be an exemplar. The second message was also updated as
follows:
a(i, k ) min{0, r (k , k ) i '{i , k } max{0, r (i ', k )}}
(7)
Meanwhile, the self-availability a(k , k ) which reflected the accumulated evidence that
point k was an exemplar for point k , was based on the positive responsibilities r (i, k ) ,
sent to candidate exemplar k from all other points. The self-availability was also updated
as follows:
a(k , k ) i '{i , k } max{0, r (i ', k )}
(8)
When updating these messages, there was a damping factor between 0 and 1 used to
avoid numerical oscillations, with a default damping value 0.5 . This message-passing
procedure was set to terminate after fixed maximal number of iterations or changes in the
messages fell below a convergence threshold (ncov=25).
After the gross parcellation of the whole brain, an average of the time series of the voxels
in the same resulting clusters was taken. Finally, new functional connectivity maps of
seed region were generated by the correlation representing the relationship between raw
seed region time series and these new whole brain time series.
In this experiment, we first averaged 25 subjects’ functional connectivity maps of the
whole brain to obtain the group whole brain functional connectivity maps and then
performed the gross clustering of the whole brain described above. By averaging the time
courses of voxels in the same clusters, we obtained new whole brain time courses in each
subjects. Then the seed region functional connectivity maps were calculated from the
correlation between seed region time courses and the new whole brain courses in each
subject. Finally, by averaging the seed region functional connectivity maps of the 25
subjects, we obtained the group seed region functional connectivity maps with the APbased feature reduction.
Raw functional Connectivity maps
Affinity Propagation
Functional connectivity
maps based on AP
Feature
Reduction
Principal Component
Analysis
Functional connectivity
maps based on PCA
Parcellation by K-means
algorithm
Comparison of three approaches
Fig. S1 Flowchart of proposed method.
Fig. S2. Illustration of examples of resulting clusters of R-SMA (K = 2 and 3). PCAall: resulting clusters using functional connectivity maps with PCA-based feature
reduction selecting all components; Raw: resulting clusters using functional
connectivity maps without feature reduction; AP: resulting clusters using functional
connectivity maps with AP-based feature reduction; K: the number of clusters
defined in K-means. The coordinate of the slice is x = 5.
Fig. S3. Illustration of examples of resulting clusters of the cingulate cortex (K = 4
and 6). PCA-all: resulting clusters using functional connectivity maps with PCAbased feature reduction selecting all components; Raw: resulting clusters using
functional connectivity maps without feature reduction; AP: resulting clusters using
functional connectivity maps with AP-based feature reduction; K: the number of
clusters defined in K-means. The coordinate of the slice is x = 5.
Table S1. One-way repeated measures ANOVA and post hoc test on the three Eigenmaps.
Repeated Measures ANOVA
P value
< 0.0001
P value summary
***
Are means signif. different? (P < 0.05)
Yes
Number of groups
3
F
18.13
R squared
0.0004013
Was the pairing significantly effective?
R squared
0.7353
F
5.559
P value
< 0.0001
P value summary
***
Is there significant matching? (P < 0.01)
Yes
Bonferroni's Multiple Comparison Test
Mean Diff.
t
Significant? P < 0.01?
Summary
R-SMA vs Cingulate Cortex
-0.00005387
4.505
Yes
***
R-SMA vs R-PFC
-0.00006833
5.713
Yes
***
Cingulate Cortex vs R-PFC
-0.00001445
1.209
No
ns
References
[1] Frey BJ, Dueck D. Clustering by passing messages between data points. Science 2007,
315: 972-976.