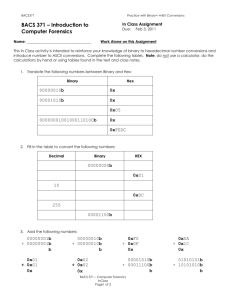
TPJ_5008_sm_TableS1-S2-FigS1-S5
advertisement

Supporting Information Table S1. Repeat motif, primer sequences, annealing temperature (Ta), size of the sequenced alleles and GenBank accession numbers of the microsatellite loci of N. attenuata Locus Primer sequence (5’-3’) Repeat motif Ta Allel Accession (°C) e size No. (bp) NA023 (TC)5 F:[6FAM]TCACTCCCTGTCTCCTTTGC 62 173 JF327433 62 88 JF327434 60 144 JF430155 60 95 JF327435 60 93 JF327436 62 241 JF327437 62 197 JF327438 62 218 JF327439 R:AATTCGAGCACCTTCAGCAG NA137 NA149 (TTC)1 F:[6FAM]ACTTTCCCCCATCTTCACCT 9 R:ACCAGGGGCTACCTGTCTTT (AAG)3 F:[6FAM]TCCAGGTCAACAAAATCAAG C R:GATGTCATTGTGCTGTCACG NA341 NA441 (TC)20 F:[HEX]AAGTCTCGTGTGGTTGCTTT (GT)9 R:AAAGGGCAATGTGTCTAGCTC (ACA)5 F:[HEX]TGGCCTCCATATGTAACCTAC C R:TCCAGACACCACTTGTGGAA NA537 (GAA)3 F:[HEX]GGTCACCGTCTTCTTCTCCA R:CCCAAATTTATCACGCAACC NA539 NA541 (AG)12 F:[HEX]GCGTGAAGCAACTAGAGAGA (CAG)3 GA (CAA)4 R: CCATCCATTGCTGCTGATAC (AAC)4 F:[HEX]CCATGTTCACTGCCTTGTTG .(ACA) R:TCCACTTTGCATTTGCACTC 3 (TAA)5 (CTT)3 Ta, annealing temperature of the primer pairs during PCR amplification reaction. 1 Table S2. Multiplex grouping and description of 16 polymorphic microsatellite markers for 20 N. attenuata native accessions. Groups Group 1 Group 2 Group 3 Marker DyeSet Repeat motif Na149a Na149b Na341a Na541a Na541b Na541c Na023a Na023b Na023c Na441a Na537a Na537b Na537c Na137a Na539a Na539b FAM FAM HEX HEX HEX HEX FAM FAM FAM HEX HEX HEX HEX FAM HEX HEX AAG AAG TC & GT AAC&ACA&TAA&CTT AAC&ACA&TAA&CTT AAC&ACA&TAA&CTT TC TC TC ACA GAA GAA GAA TTC AG&CAG&CAA AG&CAG&CAA Allele size (bp) 102-132 138-192 95-121 127-154 167-176 215-242 105-139 150-184 214-238 88-94 95-131 172-175 239-251 87-141 84-94 201-240 Na Ho He I 5 6 6 8 4 5 8 6 5 3 9 2 6 8 3 5 0.300 0.400 0.450 0.450 0.600 0.500 0.550 0.700 0.400 0.500 0.500 0.350 0.350 0.650 0.300 0.350 0.685 0.684 0.759 0.821 0.635 0.734 0.820 0.730 0.509 0.595 0.810 0.469 0.573 0.733 0.646 0.709 1.376 1.415 1.603 1.879 1.142 1.430 1.869 1.501 1.022 0.997 1.866 0.662 1.209 1.615 1.067 1.387 Na= Number of allele, Ho= Observed heterozygosity, He= Expected heterozygosity, I= Noof Heterozygotes Shannon-Weaver Information index. H o , where N is the number of N alleles; He 1 pi2 , where pi is the frequency of ith allele; I pi ln pi , where ln is the natural logarithm. 2 Figure S1. Reproducibility of nectar nicotine concentrations. Mean (±SD) nectar nicotine concentration per flower from flowers of five genotypes (Fig.1) from which nectar was collected in November 2009 (white bar; data from Fig. 1) or in October 2011 (black bar) from glasshouse grown plants. Utah wild type (UT) is fully comparable to EV (Schwachtje et al. 2008) 3 Figure S2. Allele size ranges for the 16 markers used for the genotyping of N. attenuata. Markers with non-overlapping allele size ranges amplified either by FAM labeled primers (A) or by HEX labeled primers (B) were selected for 3 different multiplex groups (shaded in different colors). 4 Figure S3. Consistent resolution of uniplex and multiplex genotyping for paternity analysis of seeds in mixed pollination. Percentage of seeds sired by 11 different pollen genotypes from mixed hand-pollinations of emasculated flowers with equal numbers of viable pollen grains in two inbred accessions of N. attenuata, UT-WT (A, C) and AZ-WT (B, D). Pollen genotypes most successful in siring seeds in UT-WT and AZ-WT are presented in exploded pie diagrams. Paternity of randomly selected 96 seeds from single capsules of UT-WT and AZ-WT plants were determined by genotyping with 16 microsatellite markers amplified separately (uniplex PCR reaction, A, B). Paternity of the same 96 samples from UT-WT and AZ-WT were determined by multiplex method of genotyping (C, D). 5 Figure S4. Consistency of microsatellite genotyping in different uniplex and multiplex PCR reactions and among biological replicates. Correlation of Shannon-Weaver Information index (I) of comparable resolution of the 16 polymorphic microsatellite markers by uniplex and multiplex genotyping technique for UT-WT (A) and AZ-WT (B) maternal genotypes. r2, correlation coefficient. 6 Figure S5. Seed paternity in an open-pollinated capsule from a single plant in a native population. Percentage of different natural paternal genotypes, detected by microsatellite genotyping, in 30 randomly selected seeds from a single, open-pollinated capsule collected from a single plant growing in a large native population in Utah. The analysis revealed that more than half of the seeds resulted from outcrossing. References Schwachtje, J., Kutschbach, S. and Baldwin, I.T. (2008) Reverse genetics in ecological research. . PLoS ONE, 3, e1543. 7
![%SYS-3-OVERRUN : Block overrun at [hex] (red zone [hex])](http://s3.studylib.net/store/data/007301636_1-ac70f3209bae6dd18e3a1bf696206cf5-300x300.png)