Chapter 1
advertisement

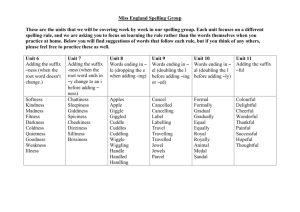
Transcription Factor Binding Sites Prediction based on Sequence Similarity Jeong Seop Sim simjs@etri.re.kr Myung Eun Lim melim@etri.re.kr Myung Geun Chung aobo@etri.re.kr Soo-Jun Park psj@etri.re.kr Sun Hee Park shp@etri.re.kr Electronics and Telecommunications Research Institute, Daejeon, Korea Abstract The availability of the whole genome sequences of human due to the Human Genome Project makes it possible to study gene function much more efficiently. We can find or predict the functions and positions of genes by analyzing the transcriptional regulation. In this paper, we propose an algorithm of predicting TF binding sites from a given set of sequences of upstream regions of genes using suffix array and local alignment algorithm. Our algorithm based on following two assumptions: i) there tends to be a common TF that binds with each of the upstream regions of the functionally related genes, ii) a TF binds with similar DNA sequences. Keywords Transcription factor, transcription factor binding sites, suffix array, local alignments Chapters’ title 2 1. Introduction Suffix trees and suffix arrays are very important index data structures in diverse applications in string processing and computational biology. Despite simplicity of suffix arrays, suffix trees have been the most fundamental index data structures in the literature because suffix arrays were inferior to suffix trees in the two aspects, construction time and search time. But recently, there have been vigorous works on suffix arrays, and suffix arrays are proved to be as powerful as suffix trees.[Kärkkäinen 2003, Kim 2003, Ko 2003,Sim 2003]. The availability of the whole genome sequences of human due to the Human Genome Project makes it possible to study gene function much more efficiently. We can find or predict the functions and positions of genes by analyzing the transcriptional regulation. In general, there are three issues in the field of transcriptional regulation, i) transcription factors (TF for short), ii) transcription factor binding sites, and iii) regulatory proteins. In this paper, we focus on the second issue and suggest an algorithm of predicting TF binding sites from a given set of sequences of upstream regions of genes (transcriptional regulation regions). For this, we need two assumptions. First, we assume that there tends to be a common TF that binds with each of the upstream regions of the functionally related genes. Our second assumption is that TF binding sites that bind with common TF have similar DNA sequences. 2. Preliminaries 2.1. Suffix array To search a pattern in a text, suffix trees and suffix arrays are widely used. The suffix tree due to McCreight [McCreight 1976] is a compacted trie of all the suffixes of a string. The suffix array due to Manber and Myers [Manber 1993] is basically a sorted list of all the suffixes of a string. To search a pattern efficiently in a suffix array, LCP (longest common prefix) information is used. An LCP array L is an array of the lengths of common prefixes of two adjacent suffixes in a suffix array SAT for a given sequence T . That is, L[i ] (1 i n 1) stores the length of common prefix of SAT [i] and SAT [i 1] where n is the length of T . For example, when a given text T abbabaababbb# where # is the smallest symbol in the alphabet, suffix array SAT of T and LCP array L are shown at Table 1. 3 Chapters’ title i SAT [i] 1 2 3 4 5 6 7 8 9 10 11 12 13 13 6 4 7 1 9 12 5 3 8 11 2 10 suffix # aababbb# abaababbb# ababbb# abbabaababbb# abbb# b# baababbb# babaababbb# babbb# bb# bbabaababbb# bbb# L[i ] 0 1 3 2 3 0 1 2 3 1 2 2 Table 1. Suffix array and LCP array 2.1. Transcription Factor Binding Sites In molecular biology, gene regulation is a hot issue and there are quite many methodologies to enable characterize gene expression patterns and profiles. Due to the huge number of cell types and organs, we cannot characterize all combinations of conditional factors by experiments. Thus, we need tools for the in silico identification of genomic signals that are related to gene regulations, for example, CoreInspector [Ohler 2001] and MCPromoter [Zhang 1998]. TRANSFAC [Matys 2003] is a database that consists of TF, TF binding sites, DNA-binding profiles. Most programs related to regulatory regions developed based on TRANSFAC. We also use TRANSFAC to test the accuracy of our algorithm. 3. 3.1. Algorithm and Results Algorithm Given a set S of DNA sequences s1 , s 2 ,, s n our algorithm first finds a common sequence above given thresholds. In general, the input sequences are set of sequences that seem to be functionally related. We use suffix arrays to find common DNA sequences in the given set of transcriptional regulation regions. First, we make a long sequence T by concatenating all the Chapters’ title 4 sequences in S . That is, T s1 #1 s2 #2 sn #n . Each # i is a special character to delimit each sequence. Now, we make a suffix array SAT of T and make LCP array L . We use Kärkkäinen and Sanders’s[Kärkkäinen 2003] algorithm to construct a suffix array of a given sequence in linear time. We are given two thresholds. One is LEN , the shortest length of the candidates of TF binding sites. The other is RTO , the frequency ratio of the candidate of TF binding sites in the given sequences. For example, if 10 sequences are given and LEN and RTO are 4 and 0.8, respectively, our algorithm finds all the sequences that are longer than 4 and appear in at least 8 sequences in the given set of sequences. See Figure 1. Figure 1. Flow of algorithm. (1) Given input sequences, our algorithm concatenates all the input sequences to make them into one sequence. (2) Build the suffix array of the concatenated sequence, and (3) make the LCP array. (4) Find sequences above given thresholds. In this example, LEN is 5 and RTO is 0.8. AGCTC is selected as a candidate. 5 Chapters’ title 3.2. Experimental Results To evaluate the accuracy of our algorithm, we obtain upstream regions and TF binding sites from EMBL release 75 and TRANSFAC [Stoesser 2003] release 3.2 to test our algorithm. We evaluated the performance of our algorithm by the measure of PPV (positive probability value, PPV = TP/(TP+FP), where TP is true positive and FP is false positive) and obtained 35.5% at best case when LEN is 4 (bp) and RTO is 0.85. See Table 2. bp % 4 5 6 7 8 9 100 30.9 18.2 0 0 0 0 95 30.9 18.2 0 0 0 0 90 33.0 14.6 0 0 0 0 85 35.3 17.5 0 0 0 0 80 29.4 13.0 15.2 25.0 0 0 75 29.6 16.9 14.3 25.0 0 0 70 30.0 7.6 13.3 25.0 0 0 65 30.2 18.1 13.3 25.0 0 0 60 27.5 15.9 10.9 10.4 8.6 7.3 55 27.6 16.7 12.1 10.0 8.2 7.3 Table 2. Positive probability value 4. Conclusion In this paper, we proposed an algorithm that predicts TF binding sites from transcription regulation regions by extracting common sequences using suffix array. Our algorithm extracts common sequences from the input sequences and output them as TF binding sites. If we search pairs of common sequences, that is, search gap allowed motifs, we think we can improve the accuracy of our algorithm. References J. Kärkkäinen and P. Sanders, Simple linear work suffix array construction, International Colloquium on Automata, Languages and Programming, LNCS 2719, 943-955, 2003. D.K. Kim, J.S. Sim, H. Park and K. Park, Linear-time construction of suffix arrays, Combinatorial Pattern Matching, LNCS 2676, 186-199, 2003. P. Ko and S. Aluru, Space efficient linear time construction of suffix arrays, Combinatorial Pattern Matching, LCNS 2676, 200-210, 2003. U. Manber and G. Myers, Suffix arrays : A new method for on-line string searches, SIAM Journal on Computing, 22, 935-938, 1993. V. Matys, E. Fricke, R. Geffers, E. Gößling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O.V. Kel-Margoulis, D.U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Chapters’ title 6 Saxel, M. Scheer, S. Thiele, and E. Wingender, TRANSFAC: transcriptional regulation, from patterns to profiles, Nucleic Acids Research, 31(1), 374-378, 2003. E.M. McCreight, A space-economical suffix tree construction algorithm, Journal of the ACM, 23, 262-272, 1976. U. Ohler, H. Niemann, G. Liao, G.M. Rubin, Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition, Bioinformatics, 17 Suppl 1, S199-206, 2001. J.S. Sim, D.K. Kim, H. Park, and K. Park, Linear-time search in suffix arrays, Australasian Workshop on Combinatorial Algorithms, 139-146, 2003. G. Stoesser, W. Baker, A. Broek, M. Garcia-Pastor, C. Kanz, T. Kulikova, R. Leinonen, Q. Lin, V. Lombard, R. Lopez, R. Mancuso, F. Nardone, P. Stoehr, M.A. Tuli, K. Tzouvara, and R. Vaughan, The EMBL ncleotide sequence database: major new developments, Nucleic Acids Research, 31(1), 17-22, 2003. M.Q. Zhang, Identification of human gene core promoters inSilico, 8(3), 319-326, 1998.