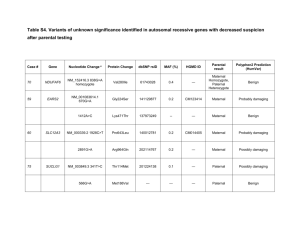
Brugada syndrome is an uncommon primary cardiac arrhythmia
advertisement

Supplemental Methods: Gene selection for screening: Like SCN5A, SCN10A (Nav1.8) encodes a TTX-resistant sodium channel and is located adjacent to SCN5A on human chromosome 3p21–22. SCN10A expression has been suggested to contribute to the late sodium current (INa) (1) and more recently the locus has been shown to act as an enhancer of SCN5A gene expression (2). TBX5 and TBX3 encode transcription factors which regulate SCN5A expression (3). HAND1 encodes a transcription factor essential for cardiac morphogenesis, and over-expression in the adult mouse leads to QRS prolongation and predisposition to ventricular arrhythmia (4). CASQ2 encodes the cardiac form of calsequestrin, a calcium-binding protein located in the lumen of the sarcoplasmic reticulum. Mutations in CASQ2 are associated with catecholaminergic polymorphic ventricular tachycardia (5). Phospholamban (PLN) is a phosphoprotein found in cardiac sarcoplasmic reticulum that is a reversible regulator of the Ca2+-ATPase activity and cardiac contractility. TKT, which encodes transketolase, an ubiquitous enzyme used in multiple metabolic pathways (6), was selected as the most likely functional candidate gene within the original QRS-associated locus (TKT/CACNA1D/PRKCD). Expression analysis in blood (7) revealed very strong cis-eQTL associations (P = 5.87×10−70) between the top QRS signal at this locus (rs4687718) and an eSNP for TKT (rs9821134) which are in moderate linkage disequilibrium (r2 = 0.47) (8). Basic procedure for ND7/23 transient transfection: The cells were cultured in Dulbecco modified Eagle’s medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 2 mmol/L L-glutamine, 100 U/mL penicillin, and 100 g/mL streptomycin at 37C under 5% CO2. After growth to ~ 70% confluence, cells were transiently transfected with 3 g of the cDNA encoding human WT or variant Nav1.8 (V1073A, A200V, or I671V) and 0.5 g of a 1 plasmid encoding green fluorescent protein (GFP) were mixed with 10 L of FuGENE6 (Omega) in 0.8 mL serum-free DMEM medium and incubated for 30 min at room temperature. The mixture was then added to the culture dish and the cells were incubated at 37C for 48 hr before the electrophysiology studies were conducted. Voltage-clamp recordings: We studied the biophysical properties of the Nav1.8 channels using whole-cell voltage-clamp protocols as previously described (1). Briefly, whole-cell currents were elicited by 100-ms pulses from -80 mV up to +50 mV in 10-mV increments from a holding potential of -120 mV. Peak inward currents were measured and plotted as a function of the test potential. Voltage dependence of Nav1.8 activation was calculated by normalizing the peak currents to the largest measured current, plotting the values as a function of voltage and fitting the data with a Boltzmann function (I/Imax = [1 + exp((V – V1/2) / k)] –1) to obtain the half maximal activation potential (V1/2) and slope factor (k). Voltage dependence of inactivation was studied using 500-ms pre-pulses from -120 to -20 mV in 10-mV increments, followed by a 100ms test pulse to -20 mV. Peak inward currents recorded at -20 mV were normalized to the peak current and plotted as a function of the pre-pulse potential. The data were then fit to a Boltzmann function to obtain V1/2 and k. The sodium window current, defined as the current range between complete inactivation and starting of activation, was also compared in this study. The pipette resistance used in this study was, on average, between 2 and 3 M, and the average access resistance was below 10 M (5-10 M). Sodium currents were recorded at room temperature using an Axopatch 200B series amplifier (Molecular Devices Corp., Sunnyvale, CA, USA). Whole-cell currents were acquired at a sampling interval of 10 kHz and filtered at 1 kHz. Pulse generation and data collection were done with Clampex 9.2. Solutions and drugs: NaV1.8 currents in transiently transfected ND7/23 cells were recorded 2 using an external potassium-free solution that contained (in mmol/L): 135 NaCl, 1.8 CaCl2, 1.0 MgCl2, 20 TEA-Cl, 10 HEPES and 10 glucose, with a pH of 7.4, adjusted with NaOH. Endogenous tetrodotoxin (TTX)-sensitive INa and L- and T-type calcium currents were eliminated with TTX 200 nM, nisoldipine 1 μM and NiCl2 200 μM, respectively, as previously reported (9, 10). Figure S1 (Supplementary data) shows the effect of these three drugs on a nontransfected ND7/23 cell. The drugs completely abolished the endogenous inward currents in ND7/23 cells. The pipette electrodes were filled with the internal solution that contained (in mmol/L): 10 NaF, 110 CsF, 20 CsCl, 5 EGTA, 10 HEPES and 5 Mg2-ATP, with a pH of 7.3 adjusted with CsOH. Supplemental data Clinical pedigree and case analysis: SCN10A E19K: The carrier of this variant presented with syncope at the age of 26 during a febrile illness and demonstrated an intermittent type 1 pattern. Ajmaline provocation was positive and atrial fibrillation, as well as polymorphic VT, were inducible at electrophysiological study. He was treated with an ICD implant. There was no family history of sudden death. No additional family was available for assessment. SCN10A R1121C: This 52 year old man was identified with the type 1 pattern following an ajmaline test due to a strong family history of premature SCD. His past medical history included atrial fibrillation. No other family members were available for assessment. TKT R148Q: This 48 year old man presented with syncope. A type 1 pattern was diagnosed on ajmaline provocation testing. Two affected relatives were available for cascade testing but did 3 not carry the putative mutation. TKT S427C: This 45 year old man presented with syncope. A type 1 pattern was diagnosed upon ajmaline provocation testing. No additional family members were available for cascade testing. TBX5 G145R: The index case was a 10 year old boy incidentally found to carry a type 1 pattern. His 38 year old mother was asymptomatic with a family history of premature SCD. She presented with a spontaneous type 1 pattern. Both were heterozygous carriers for the G145R variant. Six additional asymptomatic family members were available. None showed a type 1 pattern on standard 12 lead ECGs and five carried the G145R variant. They have not undergone testing with a sodium channel blocker. Supplemental Text References: 1. Yang T, Atack TC, Stroud DM, Zhang W, Hall L, Roden DM. Blocking SCN10A channels in heart reduces late sodium current and is antiarrhythmic. Circ. Res. 2012;111:322–332. 2. Van den Boogaard M, Smemo S, Burnicka-Turek O, et al. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J. Clin. Invest. 2014;124:1844–1852. 3. Arnolds DE, Liu F, Fahrenbach JP, et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J. Clin. Invest. 2012;122:2509–2518. 4. Breckenridge RA, Zuberi Z, Gomes J, et al. Overexpression of the transcription factor Hand1 causes predisposition towards arrhythmia in mice. J. Mol. Cell. Cardiol. 2009;47:133–141. 5. Bezzina CR, Barc J, Mizusawa Y, et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat. Genet. 2013;45:1044–1049. 6. Zhao J, Zhong C-J. A review on research progress of transketolase. Neurosci. Bull. 2009;25:94–99. 7. Dubois PCA, Trynka G, Franke L, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. 8. Sotoodehnia N, Isaacs A, de Bakker PIW, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 2010;42:1068–1076. 4 9. Holm H, Gudbjartsson DF, Arnar DO, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat. Genet. 2010;42:117–122. 10. Savio-Galimberti E, Weeke P, Muhammad R, et al. SCN10A/Nav1.8 modulation of peak and late sodium currents in patients with early onset atrial fibrillation. Cardiovasc. Res. 2014. 5 Supplemental Table S1 Gene TBX5 Exon 1(Part1) 1(Part2) 2 3 4 5 6 7 8 9(Part1) 9(Part2) TBX3 1 2 3 4 5 6 7 PLN 1 2 SCN10A 1 2 3 4 5 6 Forward primer 5’to3’ GGTATTCATTTGCCCAGAGC CCAGCCAAACGTGACAGC TTTCTCTCGTTCTCTCTCTGTCC GTGTTTTGGGGGAGTTTGG GAGGCTGCCTTAAAATACTGG CTGGTGCGTGAACTGAAGC GGGAGCAGGGTTTTATCTGG TGGCTTAATTTGCTTCTTTTGG TCTCTCACACCTGGTTCAGC TTGGCCAAATAACTGTCTCC ACTTCTCCGCTCACTTCACC AGCAGCTGCGGACTTGT AAGATTACGGGTGGTTTAT ATCCCGTTTGTTCTGCTAA ACACCCTCAAAACATTCTAGA AGGGTGTGAGGTATGTGTGT TTGGCTCATACTGGAGAT AGAGGAGAGGGATGAGATAAGC AGACTGAGACTGTGGCTAACC ATTGTATTTTTTGTTCTGAGG AAGGGCTGTTCTGACAATC CCCTTCTTGCTCATAAGCCT ATATTTATACCCACCTGTAGATCAG GTCAGAGCAGGTCATCCCTCC TCAGTCTATAGATGGCAGTGTCACTAGATTC ATGAAGAAGTGTGCTCCG Reverse primer 5’to3’ CCCAGTAAAATAAAGAGGCAACC GCCAAGTGCAAAGAGAAACC CAGACTCTGACTTTGATCTCTGC GCCACCTTTTCTTCTTCACC AACTTTTTGGGAGAAGGTTCC GAGGACAAGAGGGAGACAAGG TGCAAAAGAAAGAGCAGACG GGTTGCTGCTGGCTTACC ATACTCCTCACCCCCTCACC GCTGGAACATTCCCTCTCC TTTTTAAAATTGTGGTTTCAAGC ACCGACCAACCGACTGTTCT ACTCATGAAATGGGAAGCACT ATCCTGACTTAAAGCAGCTT ACACTTCAGAGTTGGATCCTA TTCAACTCTTCCAGGCCA ATTATTACAGCTACTAGGCCA CTGCAAAAGGAAGGGCTAAC AGAATTACCAAAGTCAGCGAA AATATTGTTTTCCTGTCTGCA GGGTGGGACCGAGAC GGGCTCTGTTGCTAACCTCTA AGACCTGGTTATTACAAAGACATATAG GGACCTGCATGTTAGCCCTGT ATAGTCTTTGCCCTGGAACCTTACAG TCCCCTGTCCCTATATGATAC Product size(bp) 478 390 297 243 248 282 280 294 390 465 474 529 398 279 212 320 839 641 234 405 458 319 452 307 338 326 Annealing temperature 59.0°C 59.0°C 59.0°C 59.0°C 59.0°C 59.0°C 59.0°C 59.0°C 59.0°C 59.0°C 59.0°C 60.0°C 58.0°C 58.0°C 50.0°C 59.0°C 60.0°C 58.0°C 61.0°C 61.0°C 54.0°C 54.0°C 51.0°C 56.4°C 55.5°C 54.0°C 6 HAND1 7 8 9 10 11 12 13 14 15(Part1) 15(Part2) 16(Part1) 16(Part2) 17 18 19 20 21 22 23 24 25 26 27(Part1) 27(Part2) 27(Part3) 1(Part1) 1(Part2) 2 TGCTGGGGGAGATTGAG ACATAAAACTCTGTGGTTGTCTCGC GGAGGCTGGTGGGTCA CCTGGCACCCCATGTC AAATGGGAGTAGGTAGAGG TAAGAGGCTGAATGATCCAAC GTCTAGAGGATGACCGCAGAA GCTGCCATTTTTATCATTGC TTTCCAGGCAAAACTTTAGA GCCATCATTGTCTTTGTC AGCTGCTAACCCGGTAGGC CAGGTCTTTGGCCATCGTAC CCTTCAAATTTAGAACTGGTC TGAACCTGCAGTGACCGTGT GGACAGGGACCACTGGTGTCATC GGGTCCTTCCCTATCAGTCTGG CATTGGAGTTCCGAATAA TGGGAGGTTCCAGGTCTTTCTCTATC CTTGGGAAGGGTCCAGTAT CACCCAGGTCCATGTCTTA TGGGCTTGGTTTGTCTATCTG TGGAAGAGGTAAGGATAGTATTTCG ATCACAGCCAACTTTGTCACG TGTGGGAGCCCAGCCGTAGG CACTGTCATTCAAAAGGCCTATCG TGGGCTGCGCGTCTCATTTT CGGCTGGAGGCGCTTGG TACGAGCCTGTTTGCCTATTT TTTTGGGGTCATTTGTTACAC GAAAGAAGAGAAACCTGTACCCATAGC ACGGTGCCCTAATTGAAGTG TGATCCACTTCCCTAAGGTGT ACAGGATGATGGCTAAAG AAGATGGTGGGGCCTAAG CCACCCCACCCGAACT CTCCCTCCCACAAGGAC AAAGGATGAGGCATATGGAT GCCATCATTGTCTTTGTC AGCTGCTAACCCGGTAGGC CCTTTTGCCCTTTGTCACTC CCTTCAAATTTAGAACTGGTC GGATCCCAGGTGAGTGTTCG AAGGTTGGGGACTTTCAGCTCAGTAG AAACTTTTCTTTGGAGTAGCCCTTGAG GGTGGTTTTGAACTCCTAA ACATAGCATCTTCTGGGAACCTAGACTCTC TGCAAGGAGGGACAGTGT GTGATGGGCTGTGAGTAGTGT ACCCTCATCCCAACGTCA CAGGGTTCTTCTAACATAAACACG TGAGGAAGGAGATGATGATGTAGG GAGAGTGCCATGGAGCGGTGC GCATTGGTGAGGCTGTAGCTGG TCTCCGCTCCTTCTTGGGT GTGTGTGCATTTTTGGAGGG GCTGCCGGCCGCGCCGCCA 477 358 401 472 491 345 400 360 401 374 425 411 398 283 542 370 387 404 257 342 366 532 546 548 411 468 560 347 54.5°C 57.2°C 55.8°C 55.4°C 55.3°C 53.8°C 57.6°C 55.0°C 54.7°C 53.7°C 58.8°C 58.1°C 55.5°C 59.0°C 58.9°C 58.1°C 54.7°C 57.3°C 55.4°C 54.1°C 56.9°C 54.7°C 57.6°C 58.8°C 57.0°C 64.0°C 60.0°C 60.0°C 7 CASQ2 TKT 1(Part1) 1(Part2) 2 3 4 5 6 7 8 9 10 11 1 2 3 4 5 6 7 8 9 10 11F & 12R 13 14 CAGCCTGTCTGCTCTCTCCT GGGCTTAATTTCCCCACATA CCCTTCCATTGATACATGAGG AGAAACCCTCAGCCAGACCT ACAACTTCCCTCCAATGCAC TTGGCTTTCTACTCTCTACACCA TGCCGGAAAACATACACACT GCAAAGGAAGCATCATCTCA TCCAGCTTCTGAGTCCCTGT TGAGCACCAAATGTCCTAAGAAG TGGTGACCTAGTTACTGTGATGG AGCTATTCAGCACGGCTATC TGAAGGCCTGGGGTGATCC CTGTGTGACTCCTCAGGACAT GTCAGTGTGAAGTAGGCACAG GCAGAGGAAGGGCTTGCTG CAGGCTCCTTGGATGGGTG GCAGACCTTTCCCTGGAAGG TAGAGGGCCAGTCTCCTGGA CTTCTCTGAAAGCAGGGCTC CTCCACAAAAGGGGTCCTAG CAGGATTCCAGCCCATGGAT GATGGCCTCTCTAGAAAGACC CCTCAGGATGTCCCACCAG CTAAGGCTGACAGAGGGCTG AAGACACCGGCTCATGGTAG ATTCTCTGCATCCTCGTTCC AAGATAGTCCGCTTATGCAAGA CCAGCTTGTCCTTTCTTTGG TGCTGAAGACCCATCCTCTT GGCAAGGGAGGATGGTTAAT GCTCTTGGCAGGTCTGAAAT GGTTTGAGTTTGGGGACTTAAT TGTGGATGGGATGTGAGAGA CCCCTGCCTATTTCACCTC CTTCTGAAAAGGCTGGGCTA ATTGCTTGCTGCCACCTT CTTCCTCGTCTAGGCATCTC AGACCACTCACCATTCCAGG CCACATGTCTGGGAAATAGGAAG CGATGGTGAAGGGTGGGTC GCACCTGCACCTGCAAAGC GGTCAGAGCTACGTAGCCAC GCTCTCCTCCCTTTCCCAG GCTCTGCCAACATGGCTGC ATCCATGGGCTGGAATCCTG TGGCCTCCTCTTCTGTTTCC AACCCCAAGACCTAGCATGC TCCTGGGGCTCAGATAGAAG GTACATCTTTGAGCACCTTTCCC 287 271 278 274 292 278 317 250 237 299 275 345 405 306 253 278 355 267 381 333 329 269 448 297 324 54.0°C 54.0°C 54.0°C 54.0°C 54.0°C 58.0°C 60.0°C 57.0°C 60.0°C 54.0°C 54.0°C 54.0°C 62.0°C 64.0°C 64.0°C 67.0°C 67.0°C 64.0°C 64.0°C 64.0°C 62.0°C 62.0°C 62.0°C 60.0°C 64.0°C 8 Supplemental Table S2: Weights used in SKAT and SKAT-O tests SNP Position on chromosome3 rs116353929 38739494 rs77804526 38739622 rs6599242 38739845 rs78425180 38740001 chr3:38740051 38740051 rs142217269 38743419 rs6790627 38748833 rs11711062 38753732 rs145032037 38753882 rs138832868 38755450 rs6771157 38763863 rs12632942 38764998 rs6795970 38766675 rs6791171 38766701 rs73062575 38766760 rs59468016 38768247 rs57326399 38768300 rs7374804 38768334 rs6800541 38774832 rs6599250 38784029 rs7630989 38793940 rs7617919 38793989 rs62244070 38798171 rs74717885 38805069 chr3:38812770 38812770 rs141207048 38835461 Minor Allele frequency 0.035 0.010 0.089 0.011 0.001 0.003 0.144 0.003 0.001 0.003 0.228 0.228 0.430 0.144 0.030 0.226 0.226 0.061 0.431 0.432 0.027 0.216 0.216 0.014 0.001 0.001 CADD score (scaled) 3.258 3.048 7.553 0.047 3.616 23.600 8.745 5.860 33 33 2.227 7.645 2.315 4.203 9.835 8.749 10.050 8.171 0.451 11.870 3.919 6.551 7.646 9.947 34.000 16.790 Allele frequency weight in SKAT test (default) 10.683 19.437 2.679 18.952 24.581 23.165 0.598 23.408 24.265 23.380 0.051 0.050 3.488E-05 0.602 12.066 0.054 0.053 5.527 3.333E-05 3.237E-05 13.020 0.072 0.073 17.682 24.561 24.176 Allele frequency in SKAT test x CADD score weights 34.804 59.245 20.231 0.891 88.886 546.700 5.230 137.171 800.743 771.536 0.113 0.383 8.075E-05 2.532 118.673 0.472 0.531 45.160 1.503E-05 3.843E-04 51.024 0.471 0.556 175.882 835.079 405.910 9 Supplemental Table S3: The classification of the 7 putative pathogenic BrS variants. GeneVariant SCN10A E19K* A200V R1121C G1299A G590R I671V TKT R148Q S427C TBX5 G145R PLN R25C *rs141810266 Predicted Classification Risk/Nonrisk Prediction Tools-Classification alleleles GERP Grantham score PolyPhen2 SIFT pathogenic pathogenic pathogenic pathogenic benign benign A/G T/C T/C C/G T/C A/G conserved conserved not conserved conserved conserved conserved benign benign damaging benign damaging benign possibly damaging probably damaging probably damaging probably damaging benign benign damaging damaging damaging damaging benign N/A pathogenic pathogenic A/G G/C conserved conserved benign damaging probably damaging probably damaging damaging damaging pathogenic A/G conserved damaging probably damaging damaging benign C/T conserved benign probably damaging benign 10 Supplemental Table S4: Collapsed common and rare variant analysis results for SCN10A Performed test MAF threshold for inclusion P-value using standard frequency weights P-value using combined frequency and CADD weights # Variants used in test # Common markers SKAT SKAT-O SKAT SKAT-O SKAT SKAT-O All All < 5% < 5% < 1% < 1% 0.2190605 0.3109835 0.2527421 0.3541483 0.5819197 0.1220259 0.4576156 0.6509294 0.492582 0.4476532 0.4465203 0.1826234 26 26 13 13 7 7 13 13 0 0 0 0 11 Supplemental Table S5: Common synonymous and non-synonymous SCN10A SNPs present in BrS patients. GeneVariant SCN10A D1739D V1697I S1622S T1570T K1441K T1131T L1092P V1073A T1064T P1045T G979G I962V K950K S509P L492L E428E I206M rsID Predicted Minor/ Genotype Frequencies Minor Allele Odds Classification Major Frequency* ratio(95%CI) alleles* Cases UK10KControls Cases Controls Hom Het Hom Hom Het Hom (minor) (major) (minor) (major) P-value rs116353929 rs77804526 rs6599242 rs78425180 rs6790627 rs6771157 rs12632942 rs6795970 rs6791171 rs73062575 rs59468016 rs57326399 rs7374804 rs7630989 rs7617919 rs62244070 rs74717885 benign benign benign benign benign benign benign benign benign benign benign benign benign benign benign benign benign 0.54 0.18 6.27x10-03 0.42 0.4 3.82x10-08 7.81x10-08 8.07x10-19 5.31x10-07 0.41 3.16x10-08 1.32x10-07 0.45 0.1 5.95x10-08 5.69x10-07 0.44 A/G T/C A/G T/C C/T C/G G/A A/G T/C T/G A/G C/T C/T G/A A/G T/C C/T 0 0 0 0 4 3 3 74 1 0 3 3 1 0 6 3 0 9 1 15 5 32 27 28 58 14 7 26 28 14 4 18 27 6 147 155 141 151 120 126 125 22 141 149 127 125 141 152 132 126 150 2 0 7 0 26 65 66 200 23 1 68 68 3 1 65 65 0 87 29 226 28 321 489 489 627 351 77 479 480 153 71 461 457 35 1190 1250 1046 1251 932 724 724 453 905 1201 731 731 1123 1207 753 757 1244 0.029 0.003 0.048 0.016 0.128 0.106 0.109 0.669 0.051 0.022 0.103 0.109 0.051 0.013 0.096 0.106 0.019 0.036 0.011 0.094 0.011 0.146 0.242 0.243 0.401 0.155 0.031 0.241 0.241 0.062 0.029 0.231 0.229 0.014 0.81(0.40-1.62) 0.28(0.04-2.07) 0.49(0.29-0.83) 1.48(0.57-3.85) 0.86(0.61-1.22) 0.37(0.25-0.54) 0.38(0.26-0.55) 3.02(2.35-3.87) 0.29(0.18-0.49) 0.72(0.33-1.58) 0.36(0.25-0.53) 0.39(0.27-0.56) 0.82(0.48-1.38) 0.44(0.16-1.22) 0.35(0.24-0.52) 0.40(0.27-0.58) 1.41(0.59-3.38) 12 13 14 15