Ribozyme
advertisement

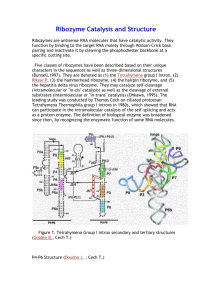
Ribozyme From Wikipedia, the free encyclopedia Jump to: navigation, search This article is about the chemical. For the rock band, see Ribozyme (band). Structure of hammerhead ribozyme A ribozyme (ribonucleic acid enzyme) is an RNA molecule that is capable of performing specific biochemical reactions, similar to the action of protein enzymes. The 1981 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. Also termed catalytic RNA, ribozymes function within the ribosome (as part of the large subunit ribosomal RNA) to link amino acids during protein synthesis, and in a variety of RNA processing reactions, including RNA splicing, viral replication, and transfer RNA biosynthesis. Examples of ribozymes include the hammerhead ribozyme, the VS ribozyme and the hairpin ribozyme. Investigators studying the origin of life have produced ribozymes in the laboratory that are capable of catalyzing their own synthesis under very specific conditions, such as an RNA polymerase ribozyme.[1] Mutagenesis and selection has been performed resulting in isolation of improved variants of the "Round-18" polymerase ribozyme from 2001. "B6.61" is able to add up to 20 nucleotides to a primer template in 24 hours, until it decomposes by cleavage of its phosphodiester bonds.[2] The "tC19Z" ribozyme can add up to 95 nucleotides with a fidelity of 0.0083 mutations/nucleotide.[3] Some ribozymes may play an important role as therapeutic agents, as enzymes which target defined RNA sequences for cleavage, as biosensors, and for applications in functional genomics and gene discovery.[4] Contents [hide] 1 Discovery 2 Activity 3 Known ribozymes 4 Artificial ribozymes 5 Applications 6 See also 7 References 8 Further reading 9 External links Discovery[edit] Schematic showing ribozyme cleavage of RNA. Before the discovery of ribozymes, enzymes, which are defined as catalytic proteins,[5] were the only known biological catalysts. In 1967, Carl Woese, Francis Crick, and Leslie Orgel were the first to suggest that RNA could act as a catalyst. This idea was based upon the discovery that RNA can form complex secondary structures.[6] The first ribozymes were discovered in the 1980s by Thomas R. Cech, who was studying RNA splicing in the ciliated protozoan Tetrahymena thermophila and Sidney Altman, who was working on the bacterial RNase P complex. These ribozymes were found in the intron of an RNA transcript, which removed itself from the transcript, as well as in the RNA component of the RNase P complex, which is involved in the maturation of pre-tRNAs. In 1989, Thomas R. Cech and Sidney Altman won the Nobel Prize in chemistry for their "discovery of catalytic properties of RNA."[7] The term ribozyme was first introduced by Kelly Kruger et al. in 1982 in a paper published in Cell.[8] It had been a firmly established belief in biology that catalysis was reserved for proteins. In retrospect, catalytic RNA makes a lot of sense. This is based on the old question regarding the origin of life: Which comes first, enzymes that do the work of the cell or nucleic acids that carry the information required to produce the enzymes? The concept of "ribonucleic acids as catalysts" circumvents this problem. RNA, in essence, can be both the chicken and the egg.[9] In the 1970s Thomas Cech, at the University of Colorado at Boulder, was studying the excision of introns in a ribosomal RNA gene in Tetrahymena thermophila. While trying to purify the enzyme responsible for splicing reaction, he found that intron could be spliced out in the absence of any added cell extract. As much as they tried, Cech and his colleagues could not identify any protein associated with the splicing reaction. After much work, Cech proposed that the intron sequence portion of the RNA could break and reform phosphodiester bonds. At about the same time, Sidney Altman, a professor at Yale University, was studying the way tRNA molecules are processed in the cell when he and his colleagues isolated an enzyme called RNase-P, which is responsible for conversion of a precursor tRNA into the active tRNA. Much to their surprise, they found that RNase-P contained RNA in addition to protein and that RNA was an essential component of the active enzyme. This was such a foreign idea that they had difficulty publishing their findings. The following year, Altman demonstrated that RNA can act as a catalyst by showing that the RNase-P RNA subunit could catalyze the cleavage of precursor tRNA into active tRNA in the absence of any protein component. Since Cech's and Altman's discovery, other investigators have discovered other examples of self-cleaving RNA or catalytic RNA molecules. Many ribozymes have either a hairpin – or hammerhead – shaped active center and a unique secondary structure that allows them to cleave other RNA molecules at specific sequences. It is now possible to make ribozymes that will specifically cleave any RNA molecule. These RNA catalysts may have pharmaceutical applications. For example, a ribozyme has been designed to cleave the RNA of HIV. If such a ribozyme were made by a cell, all incoming virus particles would have their RNA genome cleaved by the ribozyme, which would prevent infection. Activity[edit] Although most ribozymes are quite rare in the cell, their roles are sometimes essential to life. For example, the functional part of the ribosome, the molecular machine that translates RNA into proteins, is fundamentally a ribozyme, composed of RNA tertiary structural motifs that are often coordinated to metal ions such as Mg2+ as cofactors. There is no requirement for divalent cations in a five-nucleotide RNA that can catalyze trans-phenylalanation of a four-nucleotide substrate which has three base complementary sequence with the catalyst. The catalyst and substrate were devised by truncation of the C3 ribozyme.[10] RNA can also act as a hereditary molecule, which encouraged Walter Gilbert to propose that in the distant past, the cell used RNA as both the genetic material and the structural and catalytic molecule, rather than dividing these functions between DNA and protein as they are today. This hypothesis became known as the "RNA world hypothesis" of the origin of life. If ribozymes were the first molecular machines used by early life, then today's remaining ribozymes—such as the ribosome machinery—could be considered living fossils of a life based primarily on nucleic acids. A recent test-tube study of prion folding suggests that an RNA may catalyze the pathological protein conformation in the manner of a chaperone enzyme.[11] Ribozymes have been shown to be involved in the viral concatemer cleavage that precedes the packing of viral genetic material into virions.[12][13][14] Known ribozymes[edit] Naturally occurring ribozymes include: Peptidyl transferase 23S rRNA - Found in all living cells RNase P Group I and Group II introns GIR1 branching ribozyme[15] Leadzyme - Although initially created in vitro, natural examples have been found Hairpin ribozyme Hammerhead ribozyme HDV ribozyme Mammalian CPEB3 ribozyme VS ribozyme glmS ribozyme CoTC ribozyme Twister ribozyme Artificial ribozymes[edit] Since the discovery of ribozymes that exist in living organisms, there has been interest in the study of new synthetic ribozymes made in the laboratory. For example, artificially-produced self-cleaving RNAs that have good enzymatic activity have been produced. Tang and Breaker[16] isolated self-cleaving RNAs by in vitro selection of RNAs originating from random-sequence RNAs. Some of the synthetic ribozymes that were produced had novel structures, while some were similar to the naturally occurring hammerhead ribozyme. The techniques used to create artificial ribozymes involve Darwinian evolution. This approach takes advantage of RNA's dual nature as both a catalyst and an informational polymer, making it easy for an investigator to produce vast populations of RNA catalysts using polymerase enzymes. The ribozymes are mutated by reverse transcribing them with reverse transcriptase into various cDNA and amplified with mutagenic PCR. The selection parameters in these experiments often differ. One approach for selecting a ligase ribozyme involves using biotin tags, which are covalently linked to the substrate. If a molecule possesses the desired ligase activity, a streptavidin matrix can be used to recover the active molecules. Lincoln and Joyce developed an RNA enzyme system capable of self replication in about an hour. By utilizing molecular competition (in vitro evolution) of a candidate enzyme mixture, a pair of RNA enzymes emerged, in which each synthesizes the other from synthetic oligonucleotides, with no protein present.[17] Applications[edit] A type of synthetic ribozyme directed against HIV RNA called gene shears has been developed and has entered clinical testing for HIV infection.[18][19] See also[edit] Deoxyribozyme Spiegelman Monster Catalysis Enzyme RNA world hypothesis Peptide nucleic acid Nucleic acid analogues PAH world hypothesis SELEX OLE RNA References[edit] 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. Jump up ^ Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP (May 2001). "RNA-catalyzed RNA polymerization: accurate and general RNAtemplated primer extension". Science 292 (5520): 1319–1325. Bibcode:2001Sci...292.1319J. doi:10.1126/science.1060786. PMID 11358999. Jump up ^ Zaher HS, Unrau PJ (July 2007). "Selection of an improved RNA polymerase ribozyme with superior extension and fidelity". RNA 13 (7): 1017–1026. doi:10.1261/rna.548807. PMC 1894930. PMID 17586759. Jump up ^ Wochner A, Attwater J, Coulson A, Holliger P (April 2011). "Ribozyme-catalyzed transcription of an active ribozyme". Science 332 (6026): 209– 212. Bibcode:2011Sci...332..209W. doi:10.1126/science.1200752. PMID 21474753. Jump up ^ Hean J, Weinberg MS (2008). "The Hammerhead Ribozyme Revisited: New Biological Insights for the Development of Therapeutic Agents and for Reverse Genomics Applications". In Morris KL. RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Norfolk, England: Caister Academic Press. ISBN 1-904455-25-5. Jump up ^ Enzyme definition Dictionary.com Accessed 6 April 2007 Jump up ^ Carl Woese, The Genetic Code (New York: Harper and Row, 1967). Jump up ^ The Nobel Prize in Chemistry 1989 was awarded to Thomas R. Cech and Sidney Altman "for their discovery of catalytic properties of RNA". Jump up ^ Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR (November 1982). "Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena". Cell 31 (1): 147–57. doi:10.1016/0092-8674(82)90414-7. PMID 6297745. Jump up ^ Visser CM (1984). "Evolution of biocatalysis 1. Possible pregenetic-code RNA catalysts which are their own replicase". Orig. Life 14 (1–4): 291– 300. Bibcode:1984OrLi...14..291V. doi:10.1007/BF00933670. PMID 6205343. Jump up ^ Turk RM, Chumachenko NV, Yarus M (March 2010). "Multiple translational products from a five-nucleotide ribozyme". Proc. Natl. Acad. Sci. U.S.A. 107 (10): 4585–4589. Bibcode:2010PNAS..107.4585T. doi:10.1073/pnas.0912895107. PMC 2826339. PMID 20176971. Jump up ^ Supattapone S (June 2004). "Prion protein conversion in vitro". J. Mol. Med. 82 (6): 348–356. doi:10.1007/s00109-004-0534-3. PMID 15014886. Jump up ^ Herpesviruses and Chromosomal Integration. 13. 14. 15. 16. 17. 18. 19. Jump up ^ Cloning human herpes virus 6A genome into bacterial artificial chromosomes and study of DNA replication intermediates. Jump up ^ invalid reference Jump up ^ Nielsen H, Westhof E, Johansen S (2005). "An mRNA is capped by a 2', 5' lariat catalyzed by a group I-like ribozyme". Science 309 (5740): 1584– 1587. Bibcode:2005Sci...309.1584N. doi:10.1126/science.1113645. PMID 16141078. Jump up ^ Tang J, Breaker RR (May 2000). "Structural diversity of selfcleaving ribozymes". Proc. Natl. Acad. Sci. U.S.A. 97 (11): 5784–5789. Bibcode:2000PNAS...97.5784T. doi:10.1073/pnas.97.11.5784. PMC 18511. PMID 10823936. Jump up ^ Lincoln TA, Joyce GF (February 2009). "Self-sustained replication of an RNA enzyme". Science 323 (5918): 1229–1232. Bibcode:2009Sci...323.1229L. doi:10.1126/science.1167856. PMC 2652413. PMID 19131595. Jump up ^ de Feyter R, Li P (June 2000). "Technology evaluation: HIV ribozyme gene therapy, Gene Shears Pty Ltd". Curr. Opin. Mol. Ther. 2 (3): 332–5. PMID 11249628. Jump up ^ Khan AU (May 2006). "Ribozyme: a clinical tool". Clin. Chim. Acta 367 (1–2): 20–27. doi:10.1016/j.cca.2005.11.023. PMID 16426595. Further reading[edit] Astrid Sigel, Helmut Sigel and Roland K. O. Sigel, ed. (2011). Structural and catalytic roles of metal ions in RNA. Metal Ion in Life Sciences 9. Cambridge, U.K.: RSC Publishing. doi:10.1039/9781849732512. ISBN 978-1-84973-251-2. o Johnson-Buck, Alexander E.; McDowell, Sarah E.; Walter, Nils. G. "Chapter 6. Metal Ions: Supporting Actors in the Playbook of Small Ribozymes". pp. 175–196. doi:10.1039/9781849732512-00175. o Donghi, Daniela; Schnabl, Joachim. "Chapter 7. Multiple roles of metal ions in large ribozymes". pp. 197–234. doi:10.1039/9781849732512-00197. o Trappl, Krista; Polacek, Norbert. "Chapter 9. The Ribosome: A Molecular Machine Powered by RNA". pp. 253–275. doi:10.1039/978184973251200253. o Suga, Hiroaki; Futai, Kazuki; Jin, Koichiro. "Chapter 10. Metal Ion Requirements in Artificial Ribozymes that Catalyze Aminoacylation". pp. 277–297. doi:10.1039/9781849732512-00277. o Wedekind, Joseph E. "Chapter 11. Metal Ion Binding and Function in Natural and Artificial Small RNA Enzymes". pp. 299–345. doi:10.1039/9781849732512-00299. Doherty EA, Doudna JA (2001). "Ribozyme structures and mechanisms". Annu Rev Biophys Biomol Struct 30: 457–475. doi:10.1146/annurev.biophys.30.1.457. PMID 11441810. Joyce GF (2004). "Directed evolution of nucleic acid enzymes". Annu. Rev. Biochem. 73: 791–836. doi:10.1146/annurev.biochem.73.011303.073717. PMID 15189159. Ikawa Y, Tsuda K, Matsumura S, Inoue T (September 2004). "De novo synthesis and development of an RNA enzyme". Proc. Natl. Acad. Sci. U.S.A. 101 (38): 13750– 13755. Bibcode:2004PNAS..10113750I. doi:10.1073/pnas.0405886101. PMC 518828. PMID