Marker - Barley World
advertisement

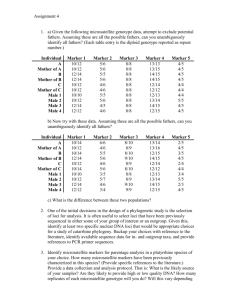
PBGMCB 620 – DNA fingerprinting Fall 2012 Homework Problem Tables 1 and 2 contain genotypic data for 10 wheat cultivars using two marker systems. Table 1 contains genotypic data using simple sequence repeat (SSR or microsatellite) markers (Roder et al., 1998; Song et al., 2005) and Table 2 contains DArt (Jaccoud et al., 2001; Wenzl et al., 2004) marker data . Evaluate the informativeness of each marker class (SSR vs. DArt) and use each dataset individually to assess genetic relatedness among some wheat cultivars described below. Perform the following calculations: 1. Calculate the number of alleles and allelic frequencies for each marker 2. Calculate the heterozygocity (Nei, 1978) and polymorphic information content (Botstein, 1980) of each marker 3. Calculate the mean heterozygocity for the microsatellite dataset 4. Calculate the mean heterozygocity for the DArT dataset 5. Which marker class was the most informative? Explain your answer by referring to your calculations. 6. Assess the genetic relatedness between the cultivars Weatherford, Madsen, ORCF101, ORCF102, and Malcolm, treating the SSR and DArt datasets separately, using the proportion-of-shared-alleles distance (Bowcock et al., 1994). 7. Assess the genetic relatedness between the cultivars Weatherford, Madsen, ORCF101, ORCF102, and Malcolm, combining the SSR and DArt datasets. Use the proportion-of-shared-alleles distance. 8. Were the estimates of genetic relatedness using different marker classes consistent with each other and with the combined dataset? Explain some reasons for consistency or lack of consistency in the analyses. Was any individual analysis consistent with the combined analysis? Explain your answer using your calculations in 6 and 7. NOTE – Show your calculations or strategy for every step. You may use any computing device or software for calculations except one that is specifically designed to do phylogenetic analysis. Table 1. Allele sizes (base-pairs) for 10 wheat cultivars screened with 10 microsatellitea (SSR) markers. Cultivar WEATHERFORD MADSEN TUBBS HILL81 MALCOLM YAMHILL HYSLOP ORCF101 ORCF102 FOOTE a barc169 119:119 119:119 119:119 119:119 119:119 119:119 116:116 119:119 119:119 116:116 barc228 172:172 179:179 172:176 172:179 176:176 172:179 179:179 176:176 172:179 172:179 barc57 236:236 224:224 224:236 230:230 236:236 224:224 230:230 224:224 224:224 215:215 gdm153 253:253 255:255 253:253 255:255 253:253 253:253 255:255 253:253 255:255 253:253 gwm11 203:203 197:197 197:197 197:197 197:197 197:197 197:197 201:201 197:197 197:197 gwm120 139:139 147:147 147:147 153:153 120:120 153:153 151:151 120:120 147:147 120:120 gwm219 182:182 182:182 182:182 190:190 147:147 182:182 190:190 147:147 147:147 147:147 gwm570 140:140 140:140 134:134 140:140 134:134 140:140 134:134 129:129 138:138 140:140 gwm95 117:117 117:117 117:117 115:115 117:117 115:115 117:117 117:117 117:117 117:117 wmc777 133:133 133:133 133:133 133:133 133:133 94:94 133:133 133:133 133:133 94:94 MIcrosatellites are described by Roder et al. (1998) and Song et al. (2005). Table 2. Allelic constitution for 10 wheat cultivars screened with 10 bi-allelic (0 = allele A and 1 = allele B) DArTa markers. MarkerName WEATHERFORD MADSEN TUBBS HILL81 MALCOLM YAMHILL HYSLOP ORCF101 ORCF102 FOOTE a wPt0950 0 0 1 0 1 0 0 1 0 1 wPt1261 1 0 0 1 0 1 0 0 1 1 wPt1741 1 1 1 1 1 0 1 0 0 0 wPt2087 0 0 1 1 1 1 0 1 1 0 wPt2781 0 1 1 1 0 1 1 0 1 1 wPt2813 1 1 1 1 0 1 1 0 0 0 DArT marker technology is described by Jaccoud et al. (2001) and Wenzl et al. (2004). wPt3524 0 1 0 1 0 1 0 1 0 1 wPt5234 1 0 0 0 1 0 1 1 0 1 wPt6975 0 1 0 0 0 1 0 0 1 1 wPt9664 1 1 1 0 1 0 1 1 1 0 References Botstein, D., White, R.L, Skolnick, M., and Davis R.W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32:314-331. Bowcock, A.M., A. Ruiz-Linares, J. Tomfohrde, E. Minch, J.R. Kidd, and L.L. CavalliSforza. 1994. High resolution of human evolutionary trees with polymorphic microsatellites 368:455-457. Jaccoud, D., K. Peng, D. Feinstein, and A. Kilian. 2001. Diversity Arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Research 29:e25-. Nei, M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583-590. Roder, M.S., V. Korzun, K. Wendehake, J. Plaschke, M.-H. Tixier, P. Leroy, and M.W. Ganal. 1998. A microsatellite map of wheat. Genetics 149:2007-2023. Song, Q.J., J.R. Shi, S. Singh, E.W. Fickus, J.M. Costa, J. Lewis, B.S. Gill, R. Ward, and P.B. Cregan. 2005. Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550-60. Wenzl, P., J. Carling, D. Kudrna, D. Jaccoud, E. Huttner, A. Kleinhofs, and A. Kilian. 2004. Diversity Arrays Technology (DArT) for whole-genome profiling of barley. Proceedings of the National Academy of Sciences USA 101:9915-9920.