HEP_24541_sm_SuppInfo
advertisement

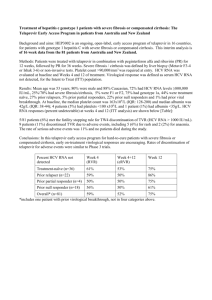
Supplementary methods Plasmid construction The HLA-B27 restricted epitope NS5B2936-2944 studied here is specific to HCV genotype 1a. Since genotype 1a based subgenomic or full-length replicons are less well established and generally exhibit inferior in vitro replication capacity as compared to genotype 1b replicons, we introduced the NS5B gene from genotype 1a (strain H77, GenBank Accession Number M67463) into a widely applied genotype 1b subgenomic replicon (pFKi341PiLucNS3-3′ET) (1) derived from the HCV strain Con1 (GenBank Accession Number AJ238799). The resulting plasmid contains the complete HCV 5′ UTR, a 63-nucleotide spacer element, the poliovirus (PV) internal ribosomal entry site (IRES), the firefly (Photinus vulgaris) luciferase gene, the IRES of the encephalomyocarditis virus, the HCV proteins NS3 to NS5A from genotype 1b strain Con1 including three cell culture–adaptive mutations (E1202G, T1280I, K1846T) that enhance viral RNA replication, the HCV protein NS5B from the genotype 1a strain H77, and the HCV 3′ UTR. Standard recombinant DNA technologies were used for all plasmid constructions. Mutations were introduced by PCR-based site-directed mutagenesis, and nucleotide sequences were confirmed by sequence analysis. Alternatively, we used a full genotype 1a based subgenomic luciferase reporter replicon, constructed analogously to the above described Con1 replicon (supplementary figure S1). It harbors the NTRs and the NS proteins 3 to 5B from strain H77 (accession number AF011751), including five previously described cell-culture adaptive mutations (Q1067R, V1655I, K1691R, K2040R, S2204I) (6), which were introduced by PCR-based mutagenesis. Nucleotide sequence was confirmed by sequence analysis. Electroporation and transient-replication assays In vitro transcription, electroporation, transient replication, and quantification of luciferase activity as a marker of HCV replication were preformed as described (2). In each assay, in addition to the mutants, the wild-type replicon as positive control, and a replication-deficient mutant of the NS5B RdRp active site (mutant GND) as negative control were run in parallel. Luciferase activity after 48 hours was normalized to the 4-hour activity (which serves as a marker for RNA transfection efficiency), and the resulting ratio was normalized to wild-type (wild-type replication = 100%). NS5B structure analyses The NS5B structure obtained from the genotype 1a strain H77 was analyzed using PyMOL (http://www.pymol.org/). Of the three entries for genotype 1a NS5B now available in the Protein Data Bank, we used 2XI3 (3) as it is the only structure with no sulfate ions potentially interfering with the side chain of R2937. Mutants were generated in PyMOL by using the rotamer closest to that of the initial residue that was consistent with the chemical environment of the mutated residue. No further optimization was performed. Structural figures were generated with PyMOL. Generation of peptide-specific T cell lines and intracellular IFN-γ staining Peptide-specific expansion of PBMC for 14 days as well as intracellular IFN-γ staining were performed as described (4). HLA-B2705 peptide binding assays Quantitative assays to measure the binding of peptides to purified HLA B*2705 class I molecules are based on the inhibition of the binding of a radiolabeled standard peptide (human 60s rL28 38-46, sequence FRYNGLIHR; 50% inhibitory concentration [IC50], 6.7 nM) and were performed as described (5). References for supplementary methods: 1. Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol 2003;77:3007-3019. 2. Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3' nontranslated region of hepatitis C virus that are important for RNA replication. J Virol 2002;76:5326-5338. 3. Harrus D, Ahmed-El-Sayed N, Simister PC, Miller S, Triconnet M, Hagedorn CH, et al. Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis. J Biol Chem 2010;285:32906-32918. 4. Neumann-Haefelin C, McKiernan S, Ward S, Viazov S, Spangenberg HC, Killinger T, et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology 2006;43:563-572. 5. Sidney J, Southwood S, Sette A. Classification of A1- and A24-supertype molecules by analysis of their MHC-peptide binding repertoires. Immunogenetics 2005;57:393-408. 6. Yi M, Lemon S. Adaptive Mutations Producing Efficient Replication of Genotype 1a Hepatitis C Virus RNA in Normal Huh7 Cells. J Virol 2004;78:7904-7915.