Towards a quantitative understanding of kinase signaling networks
advertisement

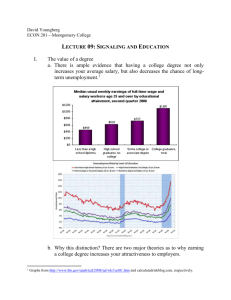
5. Stand der Wissenschaft und Technik, bisherige eigene Arbeiten, Patentlage, Wirtschaftliche Bedeutung A. Title of the project Towards a quantitative understanding of kinase signaling networks in growth and differentiation. B. Short Summary To build tissue, organs or an organism from cells complex intracellular signal transduction pathways are interconnected in a network of multifunctional, redundant and non-redundant molecules. These proteins elicit a set of phenotypic responses that subsequently impact function at the cell, tissue, and organ level. Basic principle is that the interaction of several pathways in the network can exhibit a new emergent state as a consequence of their interactions. It is this property of biological systems that makes signaling networks in growth and differentiation difficult to understand based on investigating linear pathways. Therefore, in this proposed collaborative effort we will study quantitative changes of the phosphorylation status of defined proteomes that are required for cell growth and differentiation. C. Specific Aims In response to growth-stimulatory or growth-inhibitory signals, morphogens or hormones, eukaryotic cells rely upon the timely activation and inactivation of several families of protein kinases and phosphatases1,2. These enzymes regulate progression through the cell cycle or cell cycle arrest, cell growth and differentiation, polarization and cell specification. Catastrophic human diseases such as cancer, degenerative disorders or developmental defects often result, in part, from mutations in proteins involved in these signaling pathways leading to the selective proliferation of mutant cells or lack of cell differentiation. Key molecules are the protein tyrosine (Tyr) or serine/threonine (Ser/Thr) kinases or phosphatases themselves and downstream effector molecules whose activities are controlled by phosphorylation and dephosphorylation (Fig. 1)2-4. Although many of the downstream target proteins have been identified we have only a rudimentary understanding of the assembly of signaling networks controlling growth and differentiation; and the details about how Tyr as well as Ser/Thr phosphorylation confer their unique biochemical functions to downstream effector molecules are almost entirely unknown. This is mainly due to the fact that the quantitative assessment of overall changes in protein phosphorylation has not been possible until recently. However, obtaining knowledge about the quantitative phospho-proteome is vital for a better understanding of the mechanisms controlling cell growth, differentiation and specification, and for the identification of new molecular targets against which specific therapies can be designed in human disease. Recently developed mass spectrometry-based technologies (MS) are now available that allow for the quantitative assessment of the phospho-proteome from living cells and organisms. Therefore, the major goals of the proposed project are (1) to develop and improve MS-based phospho-proteomics that can be applied to analyze the dynamic and reversible phosphorylation /dephosphorylation events that are required for cell growth and differentiation in different model systems, (2) to use these data to model and predict signaling networks required for growth and differentiation in silico, and (3) to test the obtained hypotheses in well-established model systems (such as cultured cells) and 1 organisms (including Caenorhabditis elegans, Xenopus laevis, zebrafish and Drosophila). These efforts will be combined with a novel phosphopeptide library-based proteomic screening technology5,6 that simultaneously: (1) reveals specific pSer/pThr-binding domains downstream of kinases involved in regulating cell cycle progression and cell differentiation; (2) allows determination of the optimal sequence motifs recognized by the newly-identified domain; (3) provides reagents for biophysical, cell biological, and structural studies of the function of the newly identified domain in the context of cell signaling pathways, as well as reagents for high-throughput screening of the domain for discovery of small molecule inhibitors; and (4) facilitates bioinformatics and systems biology studies to identify downstream targets of the domain that mediate cell growth and differentiation. Input: cell stimulation (Wnt, Shh, …) Dephosphorylation Phosphorylation Functional Outputs: Cell growth, differentiation Fig. 1: In response to growth-stimulatory or growth-inhibitory signals, morphogens or hormones, eukaryotic cells rely upon the timely activation and inactivation of several families of protein kinases and phosphatases. Through mediating phosphorylation/ dephosphorylation of target proteins these enzymes regulate progression through the cell cycle or cell cycle arrest, cell growth and differentiation, polarization and cell specification. D. Background and Significance Phosphorylation-Dependent Formation of Signaling Complexes. There is an increasing appreciation that many critical cell signaling events occur within large multi-molecular complexes. I.e., it has been estimated that about 500 protein complexes contain all proteins of the yeast proteome. Both the assembly and disassembly of these complexes, as well as their activity, most often appears to be regulated by specific protein phosphorylation events. Historically, the phosphorylation-dependent formation of multi-molecular signaling complexes was first elucidated for growth factor tyrosine kinase receptors3,7,8. Following binding of their respective ligands, the intracellular tyrosine kinase domains of these dimeric receptors autophosphorylate the opposite receptor in trans, generating phosphotyrosine-containing sequence motifs within the cytoplasmic non-kinase segment. These phosphotyrosine motifs, in turn, recruit the phosphoTyr-binding SH2 2 domains of other intracellular proteins, forming large multi-protein signaling complexes at the cytoplasmic tails of these receptor tyrosine kinases8. For many years it was thought that phosphoprotein-binding domains such as SH2 domains were unique to phosphoTyr, and that signaling events mediated by protein Ser/Thr phosphorylation resulted from global conformational changes in the phosphorylated protein. However, it is now more and more appreciated that Ser and Thr phosphorylation, like Tyr phosphorylation, can directly assemble signaling complexes through protein-protein interactions involving association of the pSer/pThr-containing sequence motif in one protein with a pSer/pThr-binding domain in another protein9,10. Some well-recognized phosphoserine/threonine-binding modules include 14-3-3 proteins, the proline isomerase domain and the WW domain of the mitotic regulatory protein Pin1, Forkhead Associated (FHA) domains and WD40 repeats 9,10. In many cases, these pSer/pThr-binding domains are specifically involved in regulating cell cycle progression and the response of cells to morphogens. Members of this consortium of investigators have extensive experience in the analysis of protein function and phosphorylation in cell specification and differentiation (Baumeister, Benzing, Walz, Driever, others). Canonical kinase signaling pathways that they have studied in detail include among others different MAP kinase pathways11-13, PI3 kinase signaling14 as well as canonical and non-canonical Wnt signaling pathways15 and the activation of atypical protein kinase C16. Furthermore, the groups of Timmer and Backofen have extensive experience in the data-based derivation of models for the dynamics of cellular processes. This includes methods for experimental design and estimation of parameters, testing of model and inference of system properties. In order to setup mathematical models of kinase signaling networks that are activated through morphogens and growth factors and in cell differentiation quantitative data on the phosphorylation status of the proteome will be supplemented with kinetic parameters for the individual interactions derived from the phospho-peptide library screens5,6. Thus, the foundation of this proposal is a highly interdisciplinary collaboration that brings together developmental and molecular cell biology with biochemistry, molecular medicine, biophysics and computer science. The investigators have a proven record of successful collaboration; and integration of these distinct but complementary disciplines is of central importance to our systems-level, networkbiology approach. As described above, we will attack the major goals of the proposed project (development of MS-based phospho-proteomics that can be applied to analyze the dynamic and reversible phosphorylation /dephosphorylation events; data analysis to model and predict signaling networks required for growth and differentiation in silico; experimental testing of the obtained hypotheses in wellestablished model systems) in direct interaction of these disciplines at the Center for Systems Biology (ZBSA) in Freiburg. We expect that this unique combination of methodologies will certainly allow us to successfully address kinase signaling networks controlling cell growth and to develop a better understanding of the quantitative phosphorylation events governing cell differentiation. Literature: 1. Kuriyan J, Cowburn D. Modular peptide recognition domains in eukaryotic signaling. Annu Rev Biophys Biomol Struct 1997;26:259-88. 2. Pawson T, Nash P. Protein-protein interactions define specificity in signal transduction. Genes Dev 2000;14:1027-47. 3 3. Pawson T. Protein modules and signalling networks. Nature 1995;373(6515):573-80. 4. Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science 2003;300:445-52. 5. Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing plk1 to mitotic substrates. Science 2003;299:1228-31. 6. Elia AE, Rellos P, Haire LF, et al. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the polo-box domain. Cell 2003;115(1):83-95. 7. Pawson T, Gish GD. SH2 and SH3 domains: from structure to function. Cell 1992;71(359-362). 8. Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol 2002;3(3):177-86. 9. Yaffe MB, Elia AE. Phosphoserine/threonine-binding domains. Curr Opin Cell Biol 2001;13(2):131-8. 10. Yaffe MB, Smerdon SJ. PhosphoSerine/threonine binding domains: you can't pSERious? Structure (Camb) 2001;9(3):R33-8. 11. Benzing T, Brandes R, Sellin L, et al. Upregulation of RGS7 may contribute to tumor necrosis factor-induced changes in central nervous function. Nat Med 1999;5(8):9138. 12. Benzing T, Gerke P, Hopker K, Hildebrandt F, Kim E, Walz G. Nephrocystin interacts with Pyk2, p130(Cas), and tensin and triggers phosphorylation of Pyk2. Proc Natl Acad Sci U S A 2001;98(17):9784-9. 13. Huber TB, Kottgen M, Schilling B, Walz G, Benzing T. Interaction with podocin facilitates nephrin signaling. J Biol Chem 2001;276:41543-6. 14. Huber TB, Hartleben B, Kim J, et al. Nephrin and CD2AP Associate with Phosphoinositide 3-OH Kinase and Stimulate AKT-Dependent Signaling. Mol Cell Biol 2003;23(14):4917-28. 15. Simons M, Gloy J, Ganner A, et al. Inversin, the nephronophthisis type II gene product, functions as a molecular switch between Wnt signaling pathways. Nat Genet 2005;37:537-43. 16. Benzing T. Signaling at the slit diaphragm. J Am Soc Nephrol 2004;15(6):1382-91. 4