Supplementary methods (doc 85 KB)
advertisement

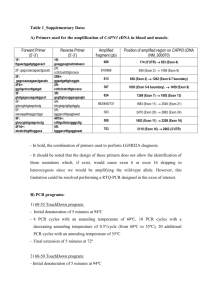
Supplementary information: methods Non-small-cell lung cancer (NSCLC) tumor tissue and immunohistochemistry Hematoxylin and eosin (H & E) staining and immunohistochemical staining (IHC) of CK7, TTF-1, and EGFR, were performed on the archival R2 lymph-node biopsy specimen with standard techniques. The cytospin slide of the cerebrospinal fluid (CSF) was stained with Papanicolou stain (Reagena, Finland). Laser microdissection (LMD) and genomic DNA extraction Tumor tissue and CSF were collected with the patient’s informed consent and in accordance with the Institutional Review Board (IRB) approved protocol at the University of Chicago. Genomic DNA was extracted from the tumor cells in the involved R2 lymph node and CSF cytospin specimen using LMD. The paraffin-embedded tumor block from the patient’s metastatic lymph node was cut onto Leica slides as 5µm thick sections, deparaffinized in xylenes and stained for H&E. Approximately 300 histopathologically identified tumor cells were then microdissected using a Leica AS LMD automated microscope with UV laser (Leica, Germany) directly into the cap of a 0.5ml PCR (polymerase chain reaction) tube (Figure 1). Genomic DNA from the tumor cells was extracted using the PicoPure DNA Extraction Kit (Arcturus, Mountain View, CA) for sequencing of the complete EGFR gene. The four tumor cells isolated from the CSF (Figure 1) were placed directly into a 15µl of a digestion buffer solution (Arcturus) and incubated at 65 °C overnight. 7.5µl of this solution was used to run a 10µl PCR reaction of exon 21 and exon 22. The entire reaction volume of the first PCR was used to run a second nested PCR reaction. The cycling conditions for the second PCR reaction were similar to the first (see supplementary information for details of PCR conditions). Leukocytes from the CSF were also microdissected to determine somatic nature of the EGFR gene mutations identified. PCR and DNA sequencing analysis EGFR PCR conditions The EGFR gene was amplified with nested PCR reactions from the DNA of laser microdissected tumor cells. The first amplification was performed using external primer pairs in a 10 µl reaction volume containing genomic DNA of 300 cells. The PCR cycling program was: 95°C for 5 min; then 94°C for 30 s, 58°C for 45 s, 72°C for 45 s for 41 cycles, with a final extension at 72°C for 10 min. 1µl of the first reaction and specified nested primers were used for the nested reaction. To avoid nonspecific PCR, a stepdown cycling program was used for the second PCR reaction: 95°C for 5 min, 94°C for 30 s, 65–58°C for 30 s, 72°C 30s for 14 cycles (annealing temperature was decreased every cycle by 0.5°C), 94°C for 30 s, 58°C for 30 s, 72°C 30 s, 72°C 10 min for 24 cycles. Aliquots of all PCR products were examined by 1.5% agarose gel electrophoresis. PCR products were purified using QIAquick PCR Purification Kit (Qiagen, Germany). Direct DNA sequencing was performed in the Core DNA Sequencing Facility at the University of Chicago (Applied Biosystems 3730XL 96-capillary automated DNA sequencer) using standard techniques. Both forward- and reverse-strand sequencing reactions were performed. EGFR genomic DNA PCR and sequencing primers are available upon request. Mutation Surveyor software program (SoftGenetics, State College, PA) was used to analyze the DNA sequencing results with subsequent manual confirmation. EGFR Expression constructs and site-directed mutagenesis Expression plasmid constructs were prepared using the full-length wild-type EGFR cDNA in pcDNA3, cloned in Xho I site. The EGFR mutations, L858R alone, E884K alone, and the double mutant L858R+E884K (identified in our patient) were introduced using the QuickChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. The plasmid vectors that resulted were sequenced to confirm the presence of the corresponding mutations introduced. In Vitro transfection and immunoblotting In vitro transfection was performed using Cos-7 cells. Transient transfection was performed as previously described using the Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Cells were plated in 10cm culture plates for transfection using 10g of plasmid vector for each of the constructs. Transfected cells were split into six-well plates, 24 hours after transfection for epidermal growth factor (EGF) stimulation studies. Cells were then starved in serum-free media for 16 hours prior to addition of the indicated concentrations of either gefitinib (gift from AstraZeneca) or erlotinib (kindly provided by Dr. Balasz Halmos). Five hours after incubation with the inhibitors, cells were stimulated with EGF as indicated (100ng/ml, 15 min) at 37oC. Whole cell lysates were then collected and separated by 7.5% SDS-PAGE for immunoblotting. The following antibodies were used for the immunoblotting: anti- phospho-EGFR [pY1068] (Cell Signaling Technology) for autophosphorylation of EGFR, anti-EGFR (Santa Cruz) for equivalent receptor expression (data not shown), and anti--actin (Sigma) as an internal loading control. Similar results were obtained from two separate immunoblotting experiments. Quantitative analysis of the phospho-EGFR signal was performed using the ImageJ software program. EGFR external primers Exon 1 Exon 2 Exon 3 Exon 4 Exon 5 Exon 6 Exon 7 Exon 8 Exon 9 Exon 10 Exon 11 Exon 12 Exon 13 Exon 14 Exon 15 Exon 16 Exon 17 GACCGGACGACAGGCCACCTCGTC (sense) GAAGAACGAAACGTCCCGTTCCTCC (antisense) GTATTATCAGTCACTAAAGCTCAC (sense) CACACTTCAAGTGGAATTCTGC (antisense) GTTGAGCACTCGTGTGCATTAGG (sense) CTCAGTGCACGTGTACTGGGTA (antisense) GTTCACTGGGCTAATTGCGGGACTCTTGTTCGCAC (sense) GGTAAATACATGCTTTTCTAGTGGTCAG (antisense) GGTCTCAAGTGATTCTACAAACCAG (sense) CCTTCACCTACTGGTTCACATCTG (antisense) CATGGTTTGACTTAGTTTGAATGTGG (sense) GGATACTAAAGATACTTTGTCACCAGG (antisense) GAACACTAGGCTGCAAAGACAGTAAC (sense) CCAAGCAAGGCAAACACATCCACC (antisense) GGAGGATGGAGCCTTTCCATCAC (sense) GAAGAGGAAGATGTGTTCCTTTGG (antisense) GAATGAAGGATGATGTGGCAGTGG (sense) GTATGTGTGAAGGAGTCACTGAAAC (antisense) GGTGAGTCACAGGTTCAGTTGC (sense) CAAAACATCAGCCATTAACGG (antisense) CCACTTACTGTTCATATAATACAGAG (sense) CATGTGAGATAGCATTTGGGAATGC (antisense) CATGACCTACCATCATTGGAAAGCAG (sense) GTAATTTCACAGTTAGGAATC (antisense) GTAGCCAGCATGTCTGTGTCAC (sense) CAGAATGCCTGTAAAGCTATAAC (antisense) GTCCTGGAGTCCCAACTCCTTGAC (sense) GGAAGTGGCTCTGATGGCCGTCCTG (antisense) CATTTGGCTTTCCCCACTCACAC (sense) GACCAAAACACCTTAAGTAACTGACTC (antisense) CCAATCCAACATCCAGACACATAG (sense) CCAGAGCCATAGAAACTTGATCAG (antisense) GAAGCTACATAGTGTCTCACTTTCC (sense) Exon 18 Exon 19 Exon 20 Exon 21 Exon 22 Exon 23 Exon 24 Exon 25 Exon 26 Exon 27 Exon 28a Exon 28b ACAACTGCTAATGGCCCGTTCTCG (antisense) CAAATGAGCTGGCAAGTGCCGTGTC(sense) GAGTTTCCCAAACACTCAGTGAAAC (antisense) GCAATATCAGCCTTAGGTGCGGCTC (sense) CATAGAAAGTGAACATTTAGGATGTG (antisense) CCATGAGTACGTATTTTGAAACTC (sense) CATATCCCCATGGCAAACTCTTGC (antisense) CTAACGTTCGCCAGCCATAAGTCC (sense) GCTGCGAGCTCACCCAGAATGTCTGG (antisense) GAGCAGCCCTGAACTCCGTCAGACTG (sense) CTCAGTACAATAGATAGACAGCAATG (antisense) CAGGACTACAGAAATGTAGGTTTC (sense) GTGCCTGCCTTAAGTAATGTGATGAC (antisense) GACTGGAAGTGTCGCATCACCAATG (sense) GGTTTAATAATGCGATCTGGGACAC (antisense) GCAGCTATAATTTAGAGAACCAAGG (sense) GGTTAAAATTGACTTCATTTCCATG (antisense) CCTAGTTGCTCTAAAACTAACG (sense) CTGTGAGGCGTGACAGCCGTGCAG (antisense) CAACCTACTAATCAGAACCAGCATC (sense) CCTTCACTGTGTCTGCAAATCTGC (antisense) GCTCCTGCTCCCTGTCATAAGTC (sense) GAAGTCCTGCTGGTAGTCAGGGTTG (antisense) CTGCAGTGGGCAACCCCGAGTATC (sense) CAGTCTGTGGGTCTAAGAGCTAATG (antisense) EGFR PCR: nested primers Exon 1 Exon 2 Exon 3 Exon 4 Exon 5 Exon 6 Exon 7 Exon 8 Exon 9 Exon 10 Exon 11 Exon 12 Exon 13 Exon 14 Exon 15 Exon 16 Exon 17 Exon 18 Exon 19 Exon 20 Exon 21 GACAGGCCACCTCGTCGGCGTC (sense) CAGCTGATCTCAAGGAAACAGG (antisense) CAGGAATGGGTGAGTCTCTGTGTG (sense) GTGGAATTCTGCCCAGGCCTTTC (antisense) CTCGTGTGCATTAGGGTTCAACTGG (sense) CCTTCTCCGAGGTGGAATTGAGTGAC (antisense) GCTAATTGCGGGACTCTTGTTCGCAC (sense) TACATGCTTTTCTAGTGGTCAG (antisense) GATTCTACAAACCAGCCAGCCAAAC (sense) CCTACTGGTTCACATCTGACCCTG (antisense) GTTTGAATGTGGTTTCGTTGGAAG (sense) CTTTGTCACCAGGCAGAGGGCAATATC (antisense) GACAGTAACTTGGGCTTTCTGAC (sense) CATCCACCCAAAGACTCTCCAAG (antisense) CCTTTCCATCACCCCTCAAGAGG (sense) GATGTGTTCCTTTGGAGGTGGCATG (antisense) GATGTGGCAGTGGCGGTTCCGGTG (sense) GGAGTCACTGAAACAAACAACAGG (antisense) GGTTCAGTTGCTTGTATAAAG (sense) ATTAACGGTAAAATTTCAGAAG (antisense) CTGTTCATATAATACAGAGTCCCTG (sense) AGAGATGCAGGAGCTCTGTGC (antisense) GCAGTTTGTAGTCAATCAAAGGTGG (sense) GTAATTTAAATGGGAATAGCCC (antisense) CCAAGGTCATGGAGCACAGG (sense) GTAAAGCTATAACAACAACCTGG (antisense) CAACTCCTTGACCATTACCTCAAG (sense) GATGGCCGTCCTGCCCACACAGG (antisense) CCACTCACACACACTAAATATTTTAAG (sense) GTAACTGACTCAAATACAAACCAC (antisense) GAGTAGTTTAGCATATATTGC (sense) GACAGTCAGAAATGCAGGAAAGC (antisense) GAAGCTACATAGTGTCTCACTTTCC (sense) CACAACTGCTAATGGCCCGTTCTCG (antisense) CAAGTGCCGTGTCCTGGCACCCAAGC (sense) CCAAACACTCAGTGAAACAAAGAG (antisense) CCTTAGGTGCGGCTCCACAGC (sense) CATTTAGGATGTGGAGATGAGC (antisense) GAAACTCAAGATCGCATTCATGC (sense) GCAAACTCTTGCTATCCCAGGAG (antisense) CAGCCATAAGTCCTCGACGTGG (sense) CATCCTCCCCTGCATGTGTTAAAC (antisense) Exon 22 Exon 23 Exon 24 Exon 25 Exon 26 Exon 27 Exon 28a Exon 28b GACGGGTCCTGGGGTGATCTGGCTC (sense) GATTACATTATCATTAGTCATTATC (antisense) GTAGGTTTCTAAACATCAAGAAAC (sense) GTGATGACATTTCTCCAGGGATGC (antisense) CATCACCAATGCCTTCTTTAAGC (sense) GCTGGAGGGTTTAATAATGCGATC (antisense) GCAAACACACAGGCACCTGCTGGC (sense) CATTTCCATGTGAGTTTCACTAGATGG (antisense) CACCTTCACAATATACCCTCCATG (sense) GACAGCCGTGCAGGGAAAAACC (antisense) GAACCAGCATCTCAAGGAGATCTC (sense) GAGCACCTGGCTTGGACACTGGAG (antisense) CCTGTCATAAGTCTCCTTGTTGAG (sense) GGTAGTCAGGGTTGTCCAGG (antisense) CGAGTATCTCAACACTGTCCAGC (sense) CTAAGAGCTAATGCGGGCATGGCTG (antisense)