100bp Std Prep
advertisement

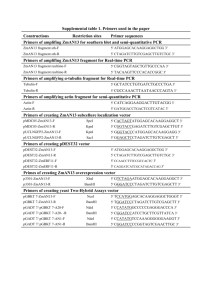
Preparation of 100 Base pair DNA Standard Reaction The set-up for multiple tubes: Reagent H2O Taq Buffer, 10X Mg2+, 25 mM dNTP mix, 10mM Forward Primer Reverse Primer Taq Polymerase, 5U/µL pBluescript Totals For 100 µL 72 10 10 4 1.5 1.5 0.5 0.5 100 x 8 Tubes 576 80 80 32 (12) (12) 4 4 800 x 2 Reactions 1152 160 160 64 (24) (24) 8 8 1600 Of which, 97 µL (reagents less primers) goes into each tube, followed by 1.5 µL each of the appropriate primers. Primers Forward ROXF ROX401F Reverse ROX104 ROX200 ROX305 ROX508 ROX597 ROX738 ROX401 ROX305 Resulting Fragment (bp) 104 200 305 508 597 738 401 800+ Only two forward primers are used. The last set is an optional addition to give an ~800bp fragment. Cycling Protocol T (°C) 94 94 Melting Time (min.) 4 1 Hold Anealing T (°C) Time (min.) 50 2 56 2 4 °C Extension T (°C) Time (min.) 72 2 72 2 Forever Repeats 33 Cleanup & Assembly Check each reaction on a gel for fragment intensity. Re-run any failed reactions. Combine all reactions in a 2 ml centrifuge tube (total is ~ 600 µL). Add 60 µL of 3M sodium acetate and 1200 µL of 100% ethanol. Incubate overnight at -20 °C. (optional) Spin for 20 minutes at 10,000 rpm. Discard supernatant. Wash pellet with 1 ml 70% ethanol. Spin for 20 minutes at 10,000 rpm. Discard supernatant. Let pellet air dry. Resuspend in 500 µL of TE buffer (sterile). Aliquot into 50 µL samples and store at -20 °C. Use ~ 2 µL/gel lane.