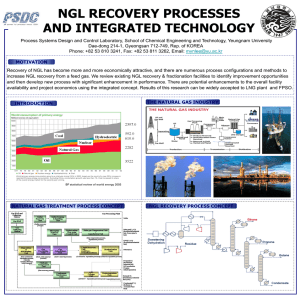
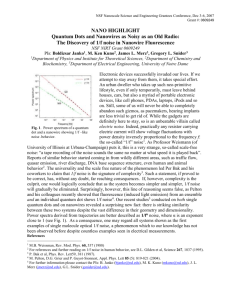
Supplementary Information
advertisement

SUPPLEMENTARY INFORMATION Spectroscopic approach for dynamic bioanalyte tracking with minimal concentration information Nicolas Spegazzini1,2,†, Ishan Barman3,4,†, Narahara Chari Dingari1,5,†, Rishikesh Pandey1, Jaqueline S. Soares1,6, Yukihiro Ozaki2 and Ramachandra Rao Dasari1,* 1 Laser Biomedical Research Center, Massachusetts Institute of Technology, Cambridge, MA 02139, USA 2 Department of Chemistry, School of Science and Technology, Kwansei Gakuin University, Sanda, Hyogo 669-1337, Japan 3 Department of Mechanical Engineering, Johns Hopkins University, Baltimore, Maryland 21218, USA 4 Department of Oncology, Johns Hopkins University, Baltimore, Maryland 21287, USA 5 Present Address: EMC Corporation, Hopkinton, MA, 01748 6 Present Address: Departamento de Física, Universidade Federal de Ouro Preto, Ouro Preto, MG 35400-000, Brazil *Correspondence: Ramachandra Rao Dasari, e-mail: rrdasari@mit.edu † Authors have made equal contributions to this work (Prepared for submission to Scientific Reports) 1 Supplementary Notes Supplementary Notes 1. Determination of the kinetic model parameters using NGL/M algorithm The gradient-based Newton-Gauss-Levenberg/Marquardt algorithm (NGL/M) is used to solve the non-linear regression problem (equation (5)) obtained by employing the iCONIC approach. In particular, the sum of squares is calculated from Q and is used as the objective function to be minimized by iteratively altering the kinetic parameter set, k. The NGL/M algorithm, which is widely used in least squares curve fitting and non-linear programming1–3, provides the means to achieve this local minimum. Briefly, the aim of the NGL/M method is to determine a vector of parameter shifts ( k ), k k new k old , that moves Qk 0 k towards zero, where k0 is the vector of initial parameter estimates. Since any gradient-based optimization method requires the calculation of a Jacobian matrix, i.e. the matrix of first partial derivatives of the residuals Q with respect to k, and as such computation in our case would lead to a three dimensional Jacobian, it is convenient to unfold Q into a long vector q (m.n×1). Then, the corresponding Jacobian JQ can be calculated by a forward finite difference, as illustrated by equation (S1): q J Q with k q qk ki qk ki ki Here, k i is the ith kinetic parameter (rate constant of the dynamic process) and ki [S1] the finite difference applied to the ith rate constant. This Jacobian matrix is used by the NGL/M algorithm to iteratively shift k towards an optimum. In particular, the vector containing the intrinsic rate constants knew, is updated from the previous vector, kold, in the following manner: k new k old J QT J Q I J QT qk 0 1 [S2] where J Q is the Jacobian matrix of Q residuals at kold, I is the identity matrix, ε is a scalar that assures that Qk new Qk old and makes the minimization algorithm immune to possible singularities of J QT J Q . 1 Convergence is estimated by the criterion of change in Q by less than 10-5 in successive iterations – conversely, the iteration process is aborted if convergence is not obtained after 1000 iterations. In order to validate successful convergence, it was further confirmed that the solution obtained did not vary when 1000 successful (i.e., not aborted) minimizations starting from randomly generated initial guesses for the rate constants were tested. This would indicate that Q has a unique well-defined minimum in the physical domain, as expected for the intrinsic rate constants. Supplementary Notes 2. Application of singular value decomposition (SVD) and suppression of baseline fluctuations In the iCONIC approach, SVD is performed on the recorded time-resolved spectral data matrix, Y, to effectively extract the critical time trace information of the analyte(s) of interest. SVD is used to decompose the data into orthonormal abstract spectra (represented by the VT matrix) and time-trace of the concentration information (represented by the U matrix). Σ is the classical diagonal matrix, where its elements are the singular values of Y. For noiseless data, the number of system 2 constituents, exhibiting detectible spectral signatures, can be obtained from ns, the number of not null elements in Σ. However, for spectral data acquired under routine experimental conditions, the singular values will not be zero beyond the expected value of ns but will tend to decay in an approximately exponential manner. Experimental time-resolved Raman spectra contain noise contributions that are both homoscedastic (where the noise is independent of the signal intensity) and heteroscedastic (where the noise is dependent on the signal intensity). The former is primarily attributable to detector noise, laser intensity fluctuations and contributions from the background. The primary source of the latter is the shot noise component, which occurs when the number of collected photons is small and hence a fluctuation in the number of detected photons causes a detectable change in the measured spectrum. The standard deviation of shot noise scales as the square root of the signal intensity. Specifically, Raman spectra with longer excitation wavelengths exhibit noise with a more homoscedastic character, since longer excitation wavelengths require higher sensitivity detectors with higher noise contributions4. Unlike shot noise, the singular value associated with baseline fluctuations can be considerably higher, leading to an overestimation of ns. To minimize this adverse impact of baseline fluctuations, first derivative-based preprocessing of the spectral dataset is pursued5. The application of the first derivative (of the spectral intensity with respect to Raman shift) completely cancels offset fluctuations and effectively minimizes more complex baselines fluctuations. Care is also taken to account for the expected noise dependence on wavenumber and time. Therefore, for our analysis, SVD is actually performed on: Yw′ = diag(wv) × der[Y] × diag(wt), (S3) where wt and wv are the inverse of the expected noise standard deviation as a function of the wavenumber and time, diag() is a diagonal matrix with diagonal elements given by the vector in the brackets, and der[ ] performs the first derivative on the corresponding matrix columns. The first derivative was performed in the Fourier domain as described in the literature6, without apodization or phase correction. As a consequence, the output of SVD becomes: Yw′ = Uw × Σ × VwT′. The normal U and V matrices are recovered by the following transformations: U = Uw × (diag(wt))−1 and VT = int[(diag(wv))−1 × VwT], where int[ ] performs the integral on the corresponding matrix columns. This process effectively pushes singular values associated with baseline fluctuations below those for genuine signals. Supplementary Information References 1. Kelley, C. T. Iterative Methods for Optimization. (SIAM frontiers in Applied Mathematics: Philadelphia, 1999). 2. Cutler, P. J., Haaland, D. M., Andries, E. & Gemperline, P. J. Methods for kinetic modeling of temporally resolved hyperspectral confocal fluorescence images. Appl. Spectrosc. 63, 153−163 (2009). 3. Cutler, P. J., Haaland, D. M. & Gemperline, P. J. Systematic method for the kinetic modeling of temporally resolved hyerspectral microscope images of fluorescently labeled cells. Appl. Spectrosc. 63, 261−270 (2009). 4. Rowlands, C. J. & Elliott, S. R. Denoising of spectra with no user input: a spline-smoothing algorithm. J. Raman Spectrosc. 42, 370−376 (2011). 5. Lórenz-Fonfría, V. A. & Kandori, H. Spectroscopic and Kinetic Evidence on How Bacteriorhodopsin Accomplishes Vectorial Proton Transport under Functional Conditions. J. Am. Chem. Soc. 131, 5891–5901 (2009). 6. Lórenz-Fonfría, V. A. & Padrós, E. Curve-fitting of Fourier manipulated spectra comprising apodization, smoothing, derivation and deconvolution. Spectrochim. Acta A 60, 2703−2710 (2004). 3 Supplementary Figure S1. Bland-Altman plot of difference in blood glucose concentrations (method – model predicted) against the mean of the two values for: (a) PLS; (b) iCONIC. 4