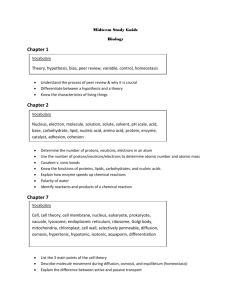
Table S4. Selected protein kinase
advertisement

Table S4. Selected protein kinase- phosphoprotein pairs suggesting functional roles for uncharacterized proteins Substrate protein Ncb2p Function * Transcriptional regulation Protein kinase (unknown Akl1p function) Pma1p, Proton pump, Pma2p regulation of pH Pheromone alpha Ste2p factor receptor Phosphatidylserin Cho1p e synthase Phospholipid binding, cell Boi2p polarity Trehalose phosphate synthase (carbohydrate metabolism, stress Tsl1p response) Polyamine Ypr156cp transport Protein kinase (unknown Ksp1p function) Protein kinase (unknown Ydr466wp function) Protein kinase (unknown Ydr466wp function) Pyruvate dehydrogenase component (pyruvate Pda1p metabolism) Protein kinase Sgv1p (transcription) Chitin synthase (cytokinesis, osmotic stress Chs2p response) Phospholuconolac Sol2p tonase (tRNA Predicted protein kinase Function * Protein kinase (unknown Akl1p function) Cellular morphogenesis, cytokinesis, endocytosis, DNA repair, cell growth, chromosome segregation, CK1 group meiosis, mitosis, nuclear ***** division Phosphopeptide ** Scansite score *** Estimated probability **** HHNSVSD 0.256 0.1 DKDSNSSITI 0.124, 0.144 0.75, 0.6 — ******* Kin4p Unknown RVSTQHE Kns1p DMYTPDTAAD 0.207 0.2 Ksp1p Unknown Protein kinase (unknown function) GTLSRRA 0.143 0.6 Ksp1p Protein kinase (unknown function) PSPTRNS 0.121 0.75 RSATRSPSA 0.146 0.6 RTSTAISRTR 0.144 0.6 NESSSTSPDE 0.144, 0.062 0.6, 0.9 SKASSEP 0.082 0.85 Cdc28p Meiosis Cyclin-dependent protein kinase, regulation of mitosis and meiosis EPSSPPP 0.02 0.99 Ygl179cp Unknown GGHSMSD 0.143 0.6 Yjl057wp Unknown AKYTSVV 0.255 0.1 Ypr106wp Unknown YRDSAHN 0.143 0.6 Ypr106wp Unknown CKSTASAAE 0.187 0.33 Protein kinase (unknown function) Protein kinase (unknown Ksp1p function) Cellular morphogenesis, cytokinesis, endocytosis, DNA repair, cell growth, chromosome segregation, meiosis, mitosis, nuclear CK1 group division Ksp1p Mek1p processing) Bre5p Unknown Bre5p Unknown Mlf3p Unknown Mlf3p Unknown Mrh1p Unknown Mrh1p Unknown Ycr023cp Unknown Ycr023cp Unknown Ydl189wp Unknown Ydl189wp Unknown Ydr090cp Unknown Yfr016cp Yfr017cp Unknown Unknown Yfr017cp Unknown Yfr024cp Unknown Yfr024cp Unknown Yhr097cp Unknown Yhr097cp Unknown Yhr132wp Unknown Cyclin-dependent protein kinase, regulation of mitosis Cdc28p and meiosis Axial budding, septin ring Kcc4p assembly Cyclin-dependent protein kinase, meiosis, regulation of Ume5p transcription (Pol II promoter) Cyclin-dependent protein kinase, meiosis, regulation of Ume5p transcription (Pol II promoter) Protein kinase (transcriptional Ctk1p regulation (Pol II promoter)) Cyclin-dependent protein kinase, regulation of mitosis Cdc28p and meiosis Actin filament organization, cell wall organization and Pkc1p biogenesis Cellular morphogenesis, cytokinesis, endocytosis, DNA repair, cell growth, chromosome segregation, meiosis, mitosis, nuclear CK1 group division Mek1p Meiosis Cyclin-dependent protein kinase, meiosis, regulation of Ume5p transcription (Pol II promoter) Actin filament organization, cell wall organization and Pkc1p biogenesis Cyclin-dependent protein kinase, meiosis, regulation of Ume5p transcription (Pol II promoter) Mek1p Meiosis cAMP-dependent protein kinase, Ras signal transduction, pseudohyphal Tpk1p growth Amino acid biosynthesis, regulation of translational Gcn2p initiation Amino acid biosynthesis, regulation of translational Gcn2p initiation Rck2p Regulation of meiosis Cellular morphogenesis, cytokinesis, endocytosis, DNA repair, cell growth, chromosome segregation, meiosis, mitosis, nuclear CK1 group division cAMP-dependent protein kinase, Ras signal transduction, pseudohyphal Tpk1p growth NASTPSSSPE 0.165 0.5 NASTPSSSPE 0.1 0.75 PATSPYV 0.084 0.85 PYVSPQQ 0.099 0.8 PAATPNL 0.145 0.6 PVASPRP 0.063 0.9 HRSSLSSLSN 0.101 0.8 HRSSLSSLSN RRASVEG 0.125 0.064 0.75 0.9 VEGSPSS 0.1 0.8 SRLSV-- — ******* TPESPKV RRRSTNY 0.02 0.127 0.99 0.75 RRSSGPM 0.19 0.33 PTNSGGSGGK 0.233 0.05 PTNSGGSGGK 0.143 ANSSTTTLD 0.077 0.6 0.85 ANSSTTTLD 0.206 0.33 RRMSSSSG 0.101 0.75 Yhr132wp Unknown *Yhr186cp Unknown Yhr186cp Unknown Yhr186cp Unknown Yml029wp Unknown Yml029wp Unknown Yml072cp Unknown Yml072cp Unknown Ymr196wp Unknown Ymr196wp Unknown Ymr295cp Unknown Ymr295cp Unknown Ynl136wp Unknown Ynl136wp Unknown Ynl156cp Unknown Ynl156cp Unknown Ynl156cp Unknown Ynl321wp Unknown Yor042wp Unknown Yor042wp Unknown Calmodulin-dependent protein kinase RRMSSSSG Actin filament organization, Pbs2p osmoregulation KAGSIQTQSR Actin filament organization, Prk1p cytokinesis ANLSTMSLVN MAPKKK, actin cytoskeleton organization and biogenesis, osmosensory signaling Ssk2p pathway ANLSTMSLVN Yak1p Cell growth and maintenance RSQSPVSFAP Cell wall organization and biogenesis, cellular Sps1p morphogenesis RSQSPVSFAP Cyclin-dependent protein kinase, regulation of mitosis Cdc28p and meiosis TSVTPRA Mitotic spindle checkpoint, Mps1p spindle pole body duplication RASSFAR Double-strand break repair, meiosis, mitotic chromosome segregation, response to Mck1p stress, sporulation SGLTPQS Amino acid biosynthesis, regulation of translational Gcn2p initiation SISSDKA Cellular morphogenesis, cytokinesis, endocytosis, DNA repair, cell growth, chromosome segregation, meiosis, mitosis, nuclear CK1 group division SSISNTSDHD Regulation of DNA replication, G1/S and G2/M transition of mitotic cell cycle, cell ion homeostasis, cell polarity, flocculation, regulation of transcription Pol CK2 group I and Pol III promoters), ****** response to DNA damage SSISNTSDHD Actin filament organization, Ark1p cytokinesis GNTSNET Double-strand break repair, meiosis, mitotic chromosome segregation, response to Mck1p stress, sporulation NETSPKR Calmodulin-dependent protein Cmk2p kinase RSVSIDSTKY Cytokinesis, regulation of exit Cdc15p from mitosis RSVSIDSTKY Calmodulin-dependent protein Cmk2p kinase RILSASSIHE Cytokinesis, regulation of exit Cdc15p from mitosis ATPSSPK MAPKKK, pseudohyphal growth, conjugation with Ste11p cellular fusion YIDTPDTETK Bub1p Mitotic spindle checkpoint YIDTPDTETK Cmk2p 0.1 0.8 0.104 0.8 0.167 0.5 0.038 0.084 0.95 0.85 0.145 0.6 0.101 0.8 0.128 0.75 0.102 0.8 0.124 0.75 0.082 0.85 0.168 0.5 0.122 0.75 0.081 0.85 0.099 0.8 0.146 0.6 0.119 0.75 0.165 0.5 0.058 0.149 0.9 0.6 Yor052cp Unknown Yor052cp Unknown Yor175cp Unknown Yor175cp Unknown Ypl247cp Unknown Yro2p Sec31p Unknown (heat shock protein homologue) Endoplasmic reticulum-Golgi transport, autophagy Receptor signalling, MAPK cascade RSSSNSSVTS Cellular morphogenesis, cytokinesis, endocytosis, DNA repair, cell growth, chromosome segregation, meiosis, mitosis, nuclear CK1 group division RSSSNSSVTS Amino acid biosynthesis, regulation of translational Gcn2p initiation MSFSGYSPKP Double-strand break repair, meiosis, mitotic chromosome segregation, response to Mck1p stress, sporulation MSFSGYSPKP Endocytosis, sphingolipid Ypk1p metabolism KRSSISF Regulation of DNA replication, G1/S and G2/M transition of mitotic cell cycle, cell ion homeostasis, cell polarity, flocculation, regulation of transcription Pol I and Pol III promoters), CK2 group response to DNA damage DVATDSE-Ykl116cp Atg1p Vesicle organization and biogenesis, autophagy RVPSLVA 0.039 0.95 0.082 0.85 0.233 0.05 0.082 0.85 0.083 0.85 — ******* 0.062 0.9 * Functional annotations based on RefSeq (Pruitt et al. 2003) ** The phosphorylated residues are underlined, and the residues not present in the phosphopeptide sequences (Ficarro et al. 2002) are shown in italic. When there is more than one phosphorylation site, the one discussed is shown in bold, unless the same protein kinase is predicted for all sites in the peptide. *** Scansite (Yaffe et al. 1999) scores were calculated as described in the Methods section. When the same protein kinase is predicted for more than one site in the peptide, the scores are given for the respective sites, starting at the N-terminus. If more than one protein kinase yields a similar score, all the possible kinases are listed. **** Probabilities were calculated as described in the Methods section (Figure 1). When the same protein kinase is predicted for more than one site in the peptide, the values are given for the respective sites, starting at the N-terminus. ***** Protein kinases Cka1p, Cka2p or Cdc7p. The predicted specificities are too similar to be distinguished. ****** Protein kinases Yck1p, Yck2p, Yck3p or Hrr25p. The predicted specificities are identical. ******* Scansite score could not be measured because the phosphorylation site is too close to the C-terminus.