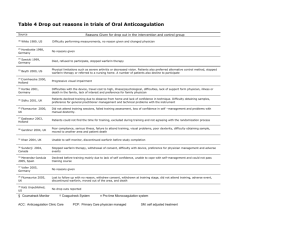
A. Treatment Protocol
advertisement

An Approach to Optimal Individualized Warfarin Treatment
through Clinical Trial Simulations
Chih-Lin Chi, Vincent A. Fusaro, Prasad Patil, Matthew A. Crawford, Charles F. Contant,
Peter J. Tonellato
Abstract—
Personalized medicine will depend on
sophisticated tools, analyses, and molecular level data and
clinical information to provide optimized treatment based on
each patient’s individual characteristics such as health
history, current health or disease status, and biochemical and
physiological makeup. We discuss an approach to integrate
clinical trial simulations with an optimization method to
produce predictions of the best individualized treatment. Our
objective is to optimize the treatment protocol by minimizing
health risk to adverse drug reactions. This approach
anticipates the era of genome-based medicine that requires
sophisticated engineering, mathematical modeling and
simulations to support best practice and clinical use of
genetic data.
Keywords— Personalize medicine, warfarin, clinical trial
simulation, optimized treatment
I.
INTRODUCTION
Individualized treatment protocols developed from
collective physicians’ experience or derived from
sophisticated clinical trials, aim to improve drug safety
and efficacy and minimize patients’ risk to serious
complications. Genetic discoveries, validated by the
statistical analysis of carefully designed clinical studies,
have produced additional genotype-dependent protocols
and algorithms. Generally, these treatment protocols result
in fewer dose-related adverse drug reactions (ADRs) when
applied to a particular population. For example, Gedge’s
warfarin dosing protocol resulted in lower ADRs when
tested in an aging population when compared to Cooper’s
protocol [5].
In addition to age, three genotypes located in two genes
(CYP2C9, VKORC1), age, race, smoking habit, and at
least 15 other dosing associated factors appear in one or
more recently published warfarin dosing algorithms. The
‘best’ protocol would reduce the likelihood of ADRs and
Manuscript received August 27, 2010. This work was supported by
U.S. National Institutes of Health under Grant R01LM010130.
Chih-Lin Chi, Vincent A. Fusaro, Prasad Patil, Matthew A.
Crawford, Peter J. Tonellato. Center for Biomedical Informatics,
Harvard Medical School. Email: {Chih-Lin_Chi, Vincent_Fusaro,
Prasad_Patil, Matthew_Crawford, Peter_Tonellato}@hms.harvard.edu
Charles F. Contant. TIMI Study Group. Phone: 617-278-0145. Fax:
617-734-7329. Address: 350 Longwood Avenue, First Floor
Boston, MA 02115, USA. Email: CCONTANT@PARTNERS.ORG
Corresponding author: Peter J. Tonellato. Joint appointment at
Center for Biomedical Informatics, Harvard Medical School and the
School of Public Health, Univ. of WI-Milwaukee. Phone: 617-4327185. Fax: 617-432-6675. Address: Center for Biomedical Informatics,
Harvard Medical School, 10 Shattuck Street, Boston, MA 02115, USA.
Email: Peter_Tonellato@hms.harvard.edu
increase the therapeutic outcome. This combination of
varying risk factors, population and individual
characteristics, large selection of treatment protocols, and
goal of reducing ADRs provides a setting to apply an
optimization method and simulations to predict the
optimal treatment protocol for each individual and attain a
central objective of personalized medicine.
In this work, we present a method and preliminary
analysis to demonstrate an approach to produce the
optimal individualized treatment protocol given a
complicated collection of individual characteristics,
genetic data, and risk to warfarin ADR. To achieve the
optimization, we will conduct a series of clinical trial
simulations and use the results of these simulations to test
an optimization methodology.
Practical clinical trials are designed to support medical
decisions by consideration of four features: (1) compare
relevant alternative interventions (2) include a diverse
population of study participants (3) recruit participants
from heterogeneous practice settings (4) use a broad range
of health outcomes [12]. Unfortunately, clinical trials are
expensive and take years to complete. In the era of
personalized medicine, the complexities of the clinical
trial increase because genetic tests and data are included.
Consequently, a new approach to treatment optimization
is needed to compliment clinical trial and other studies. In
this paper, we propose a novel approach to select the
optimal protocol based on a patient’s individual
characteristics through a method including clinical trial
simulation and optimization. Although our test case is
warfarin treatment we believe our approach is
generalizable to other personalized-medicine settings.
II. BACKGROUND
Warfarin is the most widely used anticoagulant agent in
the world, but clinical management of this drug is very
difficult because of its narrow therapeutic index [11]. For
a given individual, establishing the therapeutic dose is
difficult because the dose can vary by as much as a factor
of 10 between patients [10]. A patient’s therapeutic dose
of warfarin is influenced by many clinical (such as age)
and genetic factors (such as the CYP2C9 genotype). If
over-dosed, patients have an increased risk of bleeding
while if under-dosed patients have an increased risk of
thrombosis [1]. Several warfarin treatment protocols
were shown to reduce ADRs (i.e., risks of bleeding and
thrombosis) by reducing the time to stable therapeutic
dose. These protocols are designed to reduce the time to
initial therapeutic dose and monitor drug response to
maintain a safe and effective drug dose. (e.g., [4]).
The International normalized ratio (INR) is a standard
measurement of coagulation. For an individual, an INR
value between 2 and 3 is regarded as within the
therapeutic range [11]. Warfarin has a narrow therapeutic
window. This narrow window implies that patients may
suffer from bleeding when over-dosed (INR>4) and
thrombosis when under-dosed (low INR<2) [9, 1]. During
the course of warfarin therapy, INRs are measured several
times to help monitor the patient’s response to warfarin
dosing and guide the adjustment of dose to achieve and
maintain therapeutic response.
Adverse drug events are the primary outcome metric for
studies designed to identify improved treatment protocols.
Generally, clinical trials also use other outcomes and the
most commonly-used surrogate end point for ADRs is
time in therapeutic range (TTR), the time in the low-risk
therapeutic window over the course of the clinical trial,
often 30-60 days for warfarin trials.
Clinical trial simulations have been used by many
pharmaceutical companies to improve the efficiency of the
drug development process [8] and predict the safe dosing
of a drug prior to clinical trials. Among all simulation
approaches, pharmacokinetic and pharmacodynamic
(PK/PD) modeling and simulation has demonstrated
particular value in dose-related clinical trials because one
can use it to estimate exposure-response relationships,
predict multiple-dose profiles from a single dose, and
infer drug effectiveness and safety.
III. METHODS
We present a clinical trial simulation scenario and
individualized treatment protocols to predict the optimal
initial warfarin dosing protocol for each individual. The
method consists of four major steps: select treatment
protocol, predict the INR response for a given warfarin
dose, calculate outcome metric, and use the outcome
metric as the objective function to predict the optimal
protocol.
A. Treatment Protocol:
4
10
8
3
INR
Dose 6
(mg)
4
INR
Dose
2
2
1
1
2
3
4
Days
5
6
7
Fig 1. Adjust doses to reach the stable therapeutic dose (2≤INR≤3).
Warfarin initiation protocol continuously guide dosing adjustment
based on INR to the therapeutic dose, and predict the stable therapeutic
dose.
We use four warfarin initiation protocols to test our
approach [3, 4, 5, 10]. In general, warfarin initiation
protocols are designed to guide the establishment of the
therapeutic dose through continuous dosing adjustment
based on INR (Figure 1). The four protocols recommend a
loading dose for the first day of warfarin treatment and
then require daily testing of the patient’s INR for the first
four days. In addition, these four protocols provide
decision rules to guide the adjustment of the next dose for
the following three days based on the INR value. The
simulation then predicts the stable therapeutic dose on day
4. After four days, we continue to administer the dose for
another 10 days in order to calculate the outcome metric.
The four diverse protocols were derived and tested on
various populations. Two protocols are designed for a
normally distributed age populations [4, 10] while the
other two protocols are designed for older individuals [3,
5]. No literature compares the four protocols to provide
evidence that would help decide the “optimal” protocol.
B. Predicting the INR response
We used the Pharmacokinetic/Pharmacodynamic (PKPD model from Hamberg et al. [7] to predict the INR
response for each individual. Briefly, a PK-PD model
was derived from a set of 150 patients with median age
71, ranging from 22 to 87. Warfarin is a racemic mixture
of two enantiomers S-warfarin and R-warfarin of which Swarfarin is 3-5 times more potent and was shown to be the
dominant factor. Therefore, we only considered the PKPD effects of S-warfarin. We calculated the PK effects
using a two-compartment model with first order input and
first order elimination and the PD effects using a twochain transit compartment model. The complete
covariance matrix was not provided in the literature.
Hence, we used random normal distributions to estimate
the variability of the clearance rate, the volume in the
central compartment, and the volume in the peripheral
compartment.
To model the accumulation of warfarin dose over time
for daily doses, we used the principle of superposition.
Superpositioning does not require assumptions for PK
model or absorption kinetics, but instead assumes each
dose of the drug acts independently and that the rate and
extent of absorption and average systemic clearance are
the same for each dosing interval and that the PK model is
linear [6]. We created a table of warfarin doses over time
and summed across the rows at 24-hour time intervals to
predict the amount of warfarin remaining in the system.
C. Outcome Metric
The outcome metric used in this project is time in
therapeutic range (TTR, where 2≤INR≤3), which is the
number of days a patient stays in the therapeutic range.
We compare protocols using percentage TTR, the portion
of time staying in the therapeutic range within 14 days.
TTR represents the time to avoid bleeding and thrombosis
risks, and, thereby, is selected as the criterion to decide
the optimal protocol.
D. Predict the Optimal Individualized Warfarin Protocol
“Patients” used in the simulation framework receive
warfarin doses according to the study protocol(s) based on
calculated INR values predicted by the PK/PD model at a
given dose. A protocol decision rule system calculates the
next dose. After 14 days of simulation, the TTR is
calculated from these INRs. TTR is the percentage of the
14 days during which a patient’s INR is between 2 and 3
(the “therapeutic” range).
For each individual, the protocol with the largest
predicted TTR is identified and labeled the individualized
optimal protocol.
In the clinical trial simulation, a patient receives
warfarin treatment from four different protocols. The
PK/PD model estimates INRs based on the schedule of a
protocol, and, finally, computes the TTR for each patient.
Based on the TTR results for each protocol, the protocol
with the highest TTR was optimal.
Among four protocols, a protocol with the highest
predicted TTR is likely to translate into a highest TTR in
actual patients. Importantly, this could lead to reduced
risk of bleeding or thrombosis for a given patient. We note
that the solution space for the optimization problem is
very small, only four protocols. Consequently, an
exhaustive search solves the optimization problem.
A detail description of the general personalized
approach called a prediction and optimization-based
decision support system algorithm, is described in [2]
IV. RESULTS
Fig 2. Barplot showing the percent TTR for each of the four protocols
for a single patient.
E. Data source
To evaluate different protocols, we randomly selected
100 patients worth of data from a collection of 5700
records acquired as part of study conducted by the
International
Warfarin
Consortium
[10]
(www.pharmgkb.org). Two representative patient’s data
(Patient 1 and Patient 2) shown in Table 1 demonstrate the
individual data required for the simulation. Each patient’s
record consists of clinical (such as weight and BSA) and
genetic information (CYP2C9 and VKORC1).
Table 1
Individual patient data required for the simulation.
Parameter
Patient 1
AGE
87
HEIGHT
60
WEIGHT
112
BSA (body surface area)
1.5
GENDER
F
RACE
Asian
TARGET INR OF THE
2.5
WARFARIN TREATMENT
CYP2C9
*1/*1
VKORC1
A/A
DVT (deep vein thrombosis)
No
SMOKING
NA
AMI (amiodarone use)
No
Patient 2
57
63
183
1.9
F
AA
2.5
*3/*3
G/G
No
No
No
Fig 3. Boxplot show the distribution of TTRs for all 100 patients across
all protocols including the optimal protocol. The median value (bold
horizontal line) for each protocol is 64%, 64%, 57%, 57%, 71%
respectively. The notches on the boxplot give a visual indication of
significance between the protocols. When the notches do not overlap,
the median difference between protocols is significant.
A. Case Study of the Optimal Individualized Protocol
To clearly demonstrate the utility of simulating
protocols, we selected patient 1 as a case study (Table 1.)
and show the percent TTR for each of the four protocols
for that patient (Figure 2). The Gedge protocol is the
optimal protocol for patient 1 producing the highest TTR
(78.6%) of 11 of the 14 simulation days. Consequently,
the time out of therapeutic range is 21.4% (3 days) during
which the patient is exposed to increased risk to ADR is
the lowest compared to the other three treatment
protocols. Similar calculations are generated for each of
the 100 patients for each of the four protocols.
B. Group Results of the Optimal Individualized Protocol
Figure 3 shows the box plots of the TTR’s for all 100
patients for the four protocols and for each individual’s
optimized protocol. For these 100 randomly selected
patients, the individualized optimal protocol results in the
highest median TTR (71%, 9.94 days), mean TTR
(66.7%, 9.3 days) and lowest average time out of
therapeutic range (33.3%, 4.7 days).
The difference between average TTR (in days) between
the optimal and each of the four protocols are 1.27, 1.35,
1.32, and 1.38 days, respectively. The result indicates that
when using the individualized optimal warfarin dosing
protocol for the 100 patients, their average predicted time
in therapeutic range is slightly larger than 1 day when
compared to the non-individualized optimal warfarin
treatment protocol.
Fig 4. Parallel coordinates plot of all protocols for 100 patients. Optimal
protocols have the highest TTR from all patients. The black line in bold
represents the median of each protocol.
Figure 4 shows the predicted TTR for each of the 100
patients for each of the four treatment protocols (Fennerty,
Cooper, Gedge and Roberts from left to right) and the
optimal TTR (far right). The black line is the median
percentage of TTR of the 100 patients for each protocol.
Each protocol was the optimal protocol for
approximately 25 of the 100 randomly sampled patients
(frequency distribution in Fig 5). In some cases, the
simulations predicted that the patient’s largest TTR was
the same for more than one protocol. In those cases, we
randomly choose the “optimal” protocol.
V. DISCUSSION & CONCLUSION
Personalized medicine aims to provide the optimal
customized treatment approach for each individual based
on his or hers’ unique physiological makeup, personal and
Fig 5. Frequencies of the optimal individualized protocols for 100
patients. Each patient has an optimal protocol, and the figure shows
the frequencies from the four protocols.
family history, and genetic background. Choosing the
optimal individualized treatment amongst the many
available is a complex clinical challenge that at this time
has no obvious solution.
In this work, we demonstrate a unique computational
approach to identify the optimal protocol for an individual
patient in a post hoc manner. To do so, we randomly
selected 100 patients from a previously published warfarin
treatment study and predicted which warfarin dosing
protocol (of four) would result in the largest TTR. We
apply the prediction and optimization-based decision
support system algorithm to identify the optimal
individualized treatment protocol.
In this project, we use largest TTR as the objective to
determine the optimal protocol. TTR is the time in
therapeutic range where the therapeutic goal is to maintain
the patient’s INR between 2 and 3 during warfarin
treatment. Thus, avoiding high INR (INR > 3) and low
(INR < 2) is equally important. The method also works for
other objective functions and for combination of
objectives. For example, one might ‘weight’ certain types
of patients, or types of confounding factors. In addition,
maximal overall health benefit, minimal health risk, and
maximal cost-effectiveness can be added to the collection
of optimization criterion.
Our results show that the predicted individualized
optimal protocol has higher TTR than if each of the four
protocols were used to treat all 100 patients. If more
protocols were included, one would expect that the TTR
differences between the optimized individual treatment
protocol would result in an even higher overall TTR as
each patient is likely to be optimized over different
treatment protocols.
More protocols increase the
‘diversity’ of the protocol collection and the optimization
is likely to find a better match (lower risk) between a
given patient and a protocol.
The project shows the possibility of a new type of
decision support system that supports personalized
medicine. In this application, we predict a protocol with
the lowest risks for a patient before the patient receives
warfarin treatment, and the patient may be less likely to
suffer from risks. The system can also evaluate TTR for
every protocol as references for physicians and then
integrate the human knowledge to determine the most
appropriate protocol.
There are several limitations to our approach. First,
validation of the approach is a challenge. Our simulation
environment can predict TTR for a large collection of
protocols (in this case, four). However, there is no
opportunity to run four simultaneous or even lateral
clinical trials on the same patient to test the four (or more)
different protocols. Another limitation is the validity of
the PK/PD model. A PK/PD model is generated from a
patient population and the model optimally fits that
population. If one applies the same PK/PD model and
population parameterization to another population, the
prediction model will not be as accurate. A less serious
limitation is the protocol implementation. As discussed
earlier, we use these protocols to adjust doses for four
days. The dose at the 4th day is the predicted therapeutic
dose. In this study, we assume this predicted dose is fixed
and dose the patient for another 10 days at the same level.
We can extend our simulation to adjust dosing over the
entire 14 days but for the purpose of this demonstration
we determine to maintain the 4th day dose in an attempt to
simplify the simulation so as to focus the test on predict
TTR in a slightly simpler setting.
Next steps will include additional warfarin initiation
protocols, include warfarin maintenance protocols, test
other optimization criteria and improve the PK/PD
modeling. These additional simulations will help us
further validate our model, simulation platform and
predicted outcomes.
ACKNOWLEDGEMENT
We thank Dr. Michiyo Yamada for valuable comments.
REFERENCES
[1] JL Anderson, BD Horne, SM Stevens, AS Grove,
S Barton, ZP Nicholas, SF Kahn, HT May,
KM Samuelson, JB Muhlestein, JF Carlquist, and CoumaGen Investigators. Randomized trial of genotype-guided
versus standard warfarin dosing in patients initiating oral
anticoagulation. Circulation, 116(22):2563–2570, 2007.
[2] C.-L. Chi, W.N. Street, and M.M Ward. Building
a hospital referral expert system with a prediction and
optimization-based decision support system algorithm.
Journal of Biomedical Informatics, 41(2):371–386, 2008.
[3] M.W. Cooper and T.J. Hendra. Prospective
evaluation of a modified fennerty regimen for
anticoagulating elderly people. Age and Ageing,
27(5):655–656, 1998.
[4] A. Fennerty, J. Dolben, P. Thomas, G. Backhouse,
D.P. Bentley, I.A. Campbell, and P.A. Routledge. Flexible
induction dose regimen for warfarin and prediction of
maintenance dose. British Medical Journal,
288(28):1268–1270, 1984.
[5] J. Gedge, S. Orme, K.K. Hampton, K.S. Channer,
and T.J. Hendra. A comparison of a low-dose warfarin
induction regimen with the modified fennerty regimen in
elderly inpatients. Age and Ageing, 29(1):31–34, 2000.
[6] M. Gibaldi, , and D. Perrier. Pharmacokinetics,
Second Edition. Marcel Dekker, New York, 1982.
[7] A.K. Hamberg, M.L. Dahl, M. Barban, M.G.
Scordo, M. Wadelius, V. Pengo, R. Padrini, and E.N.
Jonsson. A pk-pd model for predicting the impact of age,
cyp2c9, and vkorc1 genotype on individualization of
warfarin therapy. Clinical pharmacology and
therapeutics, 81(4):529–538, 2007.
[8] N Holford, S.C. Ma, and B.A. Ploeger. Clinical
trial simulation: a review. Clinical Pharmacology and
Therapy, 88:166–182, 2010.
[9] A. Odén, M. Fahlén, and R.G. Hart. Optimal INR
for prevention of stroke and death in atrial fibrillation: a
critical appraisal. Thrombosis Research, 117(5):493–499,
2006.
[10] G.W. Roberts, T. Helboe, C.B. Nielsen, A.S.
Gallus, I. Jensen, D.G. Cosh, and V.S. Eaton. Assessment
of an age-adjusted warfarin initiation protocol. The Annals
of pharmacotherapy, 37(6):799–803, 2003.
[11] The International Warfarin Pharmacogenetics
Consortium. Estimation of the warfarin dose with clinical
and pharmacogenetic data. The New England Journal of
Medicine, 360(8):753–764, 2009.
[12] S.R. Tunis, D.B. Stryer, and C.M. Clancy.
Practical clinical trials: increasing the value of clinical
research for decision making in clinical and health policy.
The Journal of the American Medical Association,
290(12):1624–1632, 2003.