Genetic and molecular analyses of natural variation facilitate
advertisement

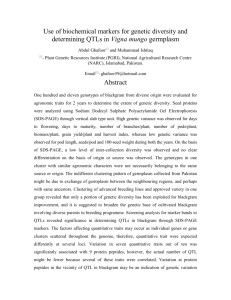
Genetic and molecular analyses of natural variation facilitate understanding of genetic control of flowering time in rice Masahiro Yano1, Atsushi Yoshimura2 and Takuji Sasaki1 1 National Institute of Agrobiological Resources, Tsukuba, Ibaraki 305-8602, Japan 2Plant Breeding Laboratory, Faculty of Agriculture, Kyushu University, Fukuoka 812-8581, Japan Recent progress in rice genome analysis has made it possible to analyze the naturally occurring allelic variation underlying complex traits, such as heading date (flowering time) of rice. We detected and characterized quantitative trait loci (QTLs) and identified genes at QTLs at the molecular level. QTLs for heading date were mapped by using several types of progeny derived from a cross between Nipponbare and Kasalath. Nine QTLs were mapped precisely as single Mendelian factors by the use of advanced backcross progeny. Nearly isogenic lines (NILs) of QTLs were also developed by marker-assisted selection and were used to identify the function of each detected QTL. Substituting Kasalath alleles of 2 QTLs into the genetic background of Nipponbare allowed us to investigate epistatic gene interactions among QTLs. We used a map-based strategy to narrow down candidate genomic regions for target QTLs. These analyses revealed 10- to 50-kb regions as candidates for the target genes. Sequencing and expression analyses were used to identify the most probable candidate genes. The function of these candidate genes was verified by a genetic complementation test. As a result, 3 QTLs—Hd1, Hd3a, and Hd6—were found to encode homologues of Arabidopsis CO, FT, and the CK2 alpha subunit, respectively. Molecular identification of other QTLs is progressing. The use of wild relatives as donor parental lines enabled us to detect additional QTLs for heading date. The analysis of natural variation has contributed greatly to our understanding of the genetic control of heading date in rice.