Supplementary Methods (doc 37K)
advertisement

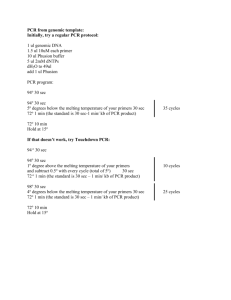
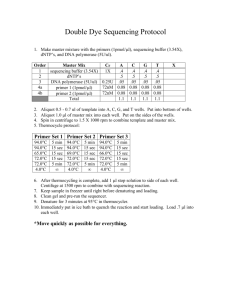
Supplementary Methods Activated Mouse ErbB2 The activated mouse ErbB2 retroviral construct (pQCXIH-ErbB2 V660E) was created using the following: Full length mouse ErbB2 cDNA (GenBank Accession BC053708) was obtained in a non-expression vector (pYX-asc) from Open Biosystems. An 81 bp insertion containing a premature stop codon (nucleotides 2259 to 2339) was removed by PCR amplification with the following primers: For 5’-CTG CTG CAG GAG ACC GAG CTG GTG GAG CCG CTG ACG-3’, and Rev 5’-CGT CAG CGG CTC CAC CAG CTC GGT CTC CTG CAG CAG-3’. This vector was then used as a template for PCR amplification of ErbB2 using a forward primer containing a 5’ extension with a Not I restriction site (For 5’-AAA GCG GCC GCG CCA CCA TGG AGC TGG CGG CCT GG-3’) and a reverse primer with a 5’ extension containing a Pac I site (Rev 5’-AAA TTA ATT AAG ACC TCA TAC TGG CAC ATC CAG GC-3’). The resulting fragment was restriction digested and cloned into the PacI/NotI sites of pQCXIH (Clontech) to yield pQCXIHErbB2. Site directed mutagenesis (QuikChange XL II, Stratagene) was performed to create a T to A transversion at nucleotide 2158. Correct full length ErbB2 and expected mutations were confirmed by DNA sequencing. Quantitation of western blots To quantify western by densitometry, developed western films were analyzed by Alpha Imager HP. Equal areas surrounding the bands were measured for bands of interest , background subtracted and integrated density(IDV) value was calculated. Average IDV’s between repeat experiments are reported in figure legends. PTPN13 mutant constructs A phosphatase inactive PTPN13 construct (pCMV5-PTPN13 C2389S) was created by site directed mutagenesis (QuikChange XL II, Stratagene) of pCMV5-PTPN13 using the following primer set: For 5’-CCA ATC ATT ACG CAC TCC AGT GCT GGC ATT GGA-3’ and Rev 5’- TCC AAT GCC AGC ACT GGA GTG CGT AAT GAT TGG-3’. pCMV5-PTPN13 T2386I, pCMV5PTPN13 P2408L, and pCMV5-PTPN13 L2423P were created by site directed mutagenesis using the following primer sets respectively: (For 5'-ACT GCC TGG CCA GAC CAT GAT ATA CCT TCT CAA CC-3' and Rev 5'-GGT TGA GAA GGT ATA TCA TGG TCT GGC CAG GCA GT-3'), ( For 5’-CAT CCA CAG ATC AGG CCT AAT CAT TAC GCA CTG CA-3’ and Rev 5’TGC AGT GCG TAA TGA TTA GGC CTG ATC TGT GGA TG-3’) and (For 5’-GGA CGT TCA GGG ACC CCG ATT TGC ATA GAT GTG-3’ and Rev 5’-CAC ATC TAT GCA AAT CGG GGT CCC TGA ACG TCC-3’). Phosphatase Activity Assay Determination of normalised phosphotyrosine phosphatase activities was done as described (Noordman et al, 2008). Briefly, COS-1 cells were transfected with pCMV5-based expression plasmids encoding N-terminally HA-tagged wild type PTPN13 or C2389S, P2408L or L2423P mutant proteins. Twenty-four hours later cells were lysed and HA-tagged proteins were immunoprecipitated using monoclonal antibody 12CA5 and subsequently released from the beads by a 30 min incubation at 4ºC in the presence of an excess HA epitope peptide (15 µg/ml; Sigma-Aldrich). Released proteins were assayed in a buffer (50 mM Hepes pH7.2; 1 mM DTT; 0.1 % BSA) containing 100microM DifMUP (Invitrogen, Breda, The Netherlands) and fluorescence emission was recorded at 460 nm at regular intervals over a 3h time period. Remaining supernatant was used for quantitative immunoblot analysis on an Odyssey infrared imaging system (LI-COR) using rabbit polyclonal antiBL-N antiserum (Wansink et al, 2004) and goat anti-rabbit secondary antibody conjugated to AlexaFluor 680 (Molecular Probes, Eugene, OR, USA). The relative PTP activity (arbitrary units) was calculated for each sample by taking the slope of the linear, steady state part of the fluorescence-over-time curves and subsequently correcting for the amount of immunoprecipitated protein. Statistical analyses (n=3) involved Student's t-test. PTPN13 Genomic Sequencing Exons 44-48 of PTPN13, were amplified from genomic DNA using PCR. The conditions for the PCR reactions are described in the supplemental methods. contained 10-40 ng DNA template, 500 nM of each forward and reverse primer, 250 uM dNTP mix, 2.5mM MgCl2, 6% DMSO, 1x PCR buffer (Invitrogen), and 2.0U Platinum Taq (Invitrogen). A touchdown Thermocycler program was used as follows: 95 ˚C for 10 min; three cycles of 94 ˚C for 30 sec, 64 ˚C for 30 sec, and 70 ˚C for 30 sec; three cycles of 94 ˚C for 30 sec, 61 ˚C for 30 sec, and 70 ˚C for 30 sec; three cycles of 94 ˚C for 30 sec, 58 ˚C for 30 sec, and 70 ˚C for 30 sec; 36 cycles of 94 ˚C for 30 sec, 57 ˚C for 30 sec, and 70 ˚C for 30 sec; and a final extension step of 70 ˚C for 5 min. PCR products were sequenced by the University of Iowa DNA Facility. Primers used for sequencing were the same as those used for PCR amplification. Sequencing results were compared to a PTPN13 reference sequence from NCBI (Accession # D21209) using SeqScape v2.5 (Applied Biosystems).