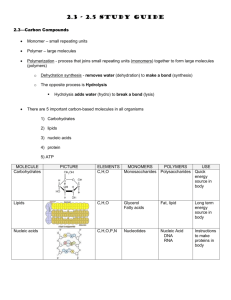
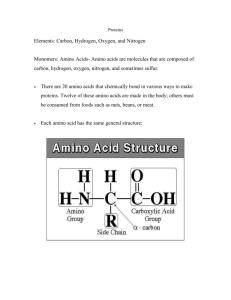
BIOL 340 Molecular Biology
advertisement

BIOL 340 Molecular Biology Lectures 2-3 Proteins Jan. 19 and 22, 2001 Reading: Lodish et al. Chap. 3 Outline: I. Structure of proteins 1. primary structure: properties of amino acids 2. secondary structure: alpha helix and beta sheet 3. tertiary structure 4. quaternary structure II. Enzymes 1. Enzyme-substrate reaction 2. Michealis-Menten kinetics 3. Variables that affect enzyme activity 4. Allosteric regulation III. Protein methods 1. Centrifugation 2. Electrophoresis 3. Liquid chromatography: gel filtration, ion exchange, affinity 4. Western Blot 5. Protein structural methods Lecture: I. Structure of proteins 1. Primary structure: sequence of amino acids linked by peptide bonds. Properties of proteins depends on the chemical groups that comprise the side chains (R groups). H | +H3N— C—COO| R R group varies for each amino acid The 20 amino acids are often grouped based on similar chemical properties (see p. 52) 1 Hydrophilic (water-soluble) Basic Acidic Lysine Aspartate Arginine Glutamate Histidine other polar amino acids Serine Threonine Asparagine Glutamine Hydrophobic Alanine Valine Isoleucine Methionine Phenylalanine Tryptophan Some amino acids can be easily remembered based on their unique properties. Amino acid(s) --containing sulfur: methionine, cystine --with H as the R group: glycine --which is an imino acid: proline --with aromatic side chains: phenylalanine, tyrosine, tryptophan --with –OH (alcohol) groups in side chain: serine, threonine How is a peptide bond formed between 2 amino acids? Carbonyl carbon from carboxylic acid group of one amino acid is linked to the amino nitrogen of the next with removal of water. Direction of protein chain growth: H2N-->-->-->COOH By convention proteins are written from the amino terminus to the carboxy terminus. 2. Secondary structure: localized arrangement of parts of the polypeptide chain in alpha helix or beta-sheet. These structures are due to a regular pattern of hydrogen bonds. Alpha helix: hydrogen bonding among residues in a protein spaced 4 apart. Shown as coil or cylinder in schematic diagrams of proteins. --Model of alpha helix in Fig. 3.6. --Carbonyl oxygen of each peptide bond is hydrogen bonded to amide nitrogen four residues toward amino terminus. 2 --Peptide backbond twists into helix with 3.6 residues/turn Beta sheet: hydrogen bonding between two chains (at least 5-8 residues at a stretch) in a protein. Can be parallel (sheets oriented in the same direction NH2-> COOH) or antiparallel. Causes the protein to form a corrogated sheet. Shown as ribbons in schematic diagrams. Concept of domains: different functional regions of proteins produced from folding of proteins in 3 dimensions. “turns” in proteins: caused by glycine and proline residues “motifs” = regions of proteins built from particular secondary structure domains examples: many in regulatory proteins, helix-loop-helix, zinc finger 3. Tertiary structure: Three dimensional folding of proteins mediated by many different types of bonds and interactions. Must be determined by X-ray crystallography or other biophysical methods. Can’t predict directly from protein sequence. However, similar regions or domains determined by studying one protein may help predict tertiary structures of proteins whose structures are not known. 4. Quaternary structure: Three dimensional structure of a multisubunit protein. II. Enzymes Alternate Reading: Campbell Chap. 6 pp. 97-104 Enzymes are catalysts, substances that speed up the rate of a reaction. They are nearly always proteins, although there are some enzymes such as the one that makes proteins (peptidyl transferase) which are catalytic RNAs. Enzymes like other catalysts are not themselves used up by the reaction. How do enzymes speed up the rate of the reaction? Reactions involve the breaking and reforming of chemical bonds. The breaking of bonds needs energy to get started even if the overall process is energetically favorable. The energy needed to get started is called the “activation energy”. Figure 6-10 Campbell The activation energy represents the uphill portion of the graph. 3 As the reactants absorb energy, they become unstable. This is the “transition state”. As new bonds are formed, energy is released into the surroundings. This is the downhill portion of the curve which indicates a loss of free energy by the products. The difference in free energy of the products vs. the reactants is G for the reaction which is negative for an exergonic reaction. To overcome the activation energy requires energy input. For some chemical reactions, this can be accomplished by heating. Cells can be damaged by heat, therefore they use enzyme catalysts to lower the activation energy. An enzyme cannot change the G for a reaction. They speed up reactions that would have occurred anyway, but perhaps at an extremely slow rate. Fig. 6.10 shows how enzymes lower the EA without affecting G for the reaction. 2. Substrate binding Enzyme catalyzed reactions can be summarized as follows: substrate enzyme ----> products substrate = reactant(s) acted on by an enzyme Example sucrose + H2O sucrase ----> glucose substrates + fructose products Enzymes usually are highly specific for their substrates. The region of the enzyme responsible for binding to substrate is called the “active site”. The active site is typically a pocket or groove created by some of the amino acids of the enzyme. Fig. 6-11 shows binding of the substrate (glucose) to the groove that makes up the active site of hexokinase. Models that explain different aspects of substrate binding 1) lock and key --good for explaining specificity 4 --doesn’t explain why substrates that resemble the transition state are the best fit 2) induced fit Substrate induces a conformational change that creates better fit of enzyme to substrate and places bonds in key positions to be acted on by the substrate. Key features of enzyme/substrate interations: --Substrate is bound to the enzyme by many weak interactions (hydrogen bonds, ionic bonds, hydrophobic interactions). --Catalysis is begun by amino acids making up the active site of the enzyme; R groups are usually crucial in catalysis. --Enzymes can catalyze thousands of reactions per second. --The more substrate available, the more rapid the reaction, until the enzyme is saturated by substrate. How do we measure the rate of an enzyme catalyzed reaction? Mix --enzyme --substrate (vary) --buffer (maintain correct pH for enzyme activity) Measure amount of product produced. 3. Factors that affect enzyme activity a. Temperature: Most enzymes have optimal activity at body temperature (37C). Enzymes from thermophilic organisms may have optimal activity at 70C. b. pH: Most enzymes have optimal activity around pH 7. Pepsin, digestive enzyme of stomach works best in the acid environment of the stomach, pH 2.0. c. cofactors: Ions or coenzymes that are required by the enzyme for catalytic activity. d. inhibitors: Chemicals that slow or stop the action of enzymes. 5 competitive inhibitors: Bind to active site, prevent access of substrate to active site. noncompetitive inhibitors: Bind to enzyme somewhere other than the active site. 4. Enzyme regulation Allows cell to respond to environmental signals and slow down or speed up steps in metabolism. allosteric regulation: Binding of a small molecule to site other that the active site which changes the enzyme conformation. feedback inhibition: When end product in reaction signals to enzyme at beginning of pathway to slow down. A ---> B ---> C ---> D D is a feed back inhibitor of enzyme that converts A to B Enzymes are mostly protein catalysts that speed up the rate of a chemical reaction in cells. Enzymes must be in their proper conformation to bind substrate and catalyze reactions. Changes in pH or temperature or presence of chemical inhibitors can inactivate enzymes. III. Protein Methods 1. Centrifugation: sediment particles of differerent masses or densities using centrifugal force. Differential centrifugation: separate particles of different masses; the particle with more mass forms a pellet at the bottom of the tube. Density gradient centrifugation: separate molecules of different mass but similar size and density. 2. Electrophoresis: separate molecules in an electric field SDS-polyacrylamide gel electrophoresis: --used for separation of proteins. --gel formed by forming a chain of monomers (acrylamide) with occaisional cross-links (bis-acrylamide) --SDS denatures proteins, causes proteins to have similar (-) charges --separates proteins according to size; small proteins migrate farther. 2-D gel electrophoresis: --use to separate proteins that may have the same mass. 6 --first separate based on charge by isoelectric focusing. --use SDS-PAGE to separate in the second dimension based on size. --generate a series of spots each representing a unique protein 3. Liquid chromatography: Separate molecules in solution based on their interactions with a solid surface. A. gel filtration: separate proteins based on different mass B. ion exchange: separate proteins based on charge C. affinity chromatography: separate proteins based on affinity of protein for another molecule 4. Western Blot (Immunoblot): Use of SDS-PAGE to separate proteins followed by electrophoretic transfer of proteins to membrane. Specific protein of interest detected using an antibody. 5. Protein structural methods A. Primary structure: i. amino acid sequencing by “Edman degradation” amino terminus --> carboxy terminus ii. translate gene sequence B. 3-D structure Most important method is X-ray crystallography. 7 8