here
advertisement

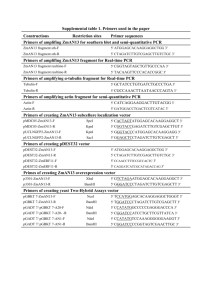
1 Supplementary material 2 Construction of R. solanacearum mutants 3 Construction of a eps mutant. The disruption of an eps gene in R. solanacearum 4 RS1002 was performed using the marker-exchange plasmid pK18mobsacB (Schäfer et 5 al., 1994). A 2.4 kb fragment including the epsP gene was amplified by polymerase 6 chain 7 (5'-CGTAAGCATTCCGAGTATCC-3') 8 (5'-TTGCGCTTGATGTTCTGGGC-3') and RS1002 genomic DNA as the template. 9 PCR was performed using the GC-RICH PCR system (Roche). The PCR fragment was 10 cloned into pK18mobsacB to yield pRS183. Next, deletion was carried out in the epsP 11 gene by excluding a 0.4 kb EcoRV-NruI fragment from pRS189. Using pRS189, the 12 EPS- strain RS1085 was constructed from RS1002 by the marker-exchange method 13 (Schäfer et al., 1994). reaction (PCR) using the primer and set of P26 P29 14 15 Construction of (hrpB-hrpD) mutants. First, a hrp gene cluster was cloned from R. 16 solanacearum RS1002. A part of the popA gene was amplified by PCR using primers P1 17 (5'-AGGTGCCCGCTCGTTAC-3') and P2 (5'-CCATCGCCATTACATCGGCT-3') and 18 RS1002 genomic DNA. The 1.1 kb fragment was cloned into a suicide vector pARO191 19 (Parke, 1990) to yield pINT101. pINT101 was integrated into the genome of RS1002 20 through conjugation with S17-1. Genomic DNA was purified from a transconjugant and 21 the integrated plasmid was rescued to include the surrounding hrp region. The partial 22 sequencing of the rescued plasmid pRS103 revealed that the hrp gene cluster of RS1002 23 has the same gene organization as that of GMI1000 (Van Gijsegem et al., 1995). A 4.0 24 kb EcoRI fragment containing the hrpB-hrcT-hrpD-hrcN-hrpF region from pRS103 was 25 subcloned into pK18mobsacB, and a 1.3 kb XhoI fragment including the 1 1 hrpB-hrcT-hrpD region was replaced with a gentamycin resistance (Gmr) gene cassette, 2 yielding pRS121. Using pRS121, (hrpB-hrpD) strains were constructed by the 3 marker-exchange method. 4 5 Construction of hpaB mutants. A 0.9 kb fragment upstream of hpaB was PCR 6 amplified 7 (5'-GGGAAGCTTGTGTGCAACGGACTTATCCC-3') 8 (5'-GGGGGATCCGTGTGCAACGGACTTATCCC-3') with HindIII (underlined) and 9 BamHI (double underlined) restriction sites, respectively. Similarly, a 1.0 kb fragment from RS1002 genomic DNA using primers and P691 P694 10 downstream 11 (5'-GGGGGATCCAGACCACCGACGACATGTT-3') 12 (5'-GGGGGTACCAACAGGTTCATGAACCGCGAGC-3') with BamHI (underlined) 13 and KpnI (double underlined) sites, respectively. The two fragments were tandemly 14 inserted into pK18mobsacB. The resulting pRS769 (Fig. 3) contains a 340 bp deletion in 15 hpaB. hpaB strains were constructed by marker exchange using pRS769. Primers P691 16 and P698 were used for the selection of hpaB mutants by PCR. of hpaB was amplified using primers and P695 P698 17 18 Construction of hrpY mutants. A 0.8 kb EcoRI-PstI fragment containing the hrpY gene 19 was cloned from pRS103 into pHSG398, yielding pRS771. pRS771 was digested with 20 SalI and EcoO109I, treated by Klenow and self-ligated to result in a 160 bp deletion in 21 hrpY. The EcoRI-PstI fragment containing hrpY was subcloned into pK18mobsacB to 22 yield pRS828. hrpY strains were constructed by marker exchange using pRS828. 23 Primers P691 and P694 were used for the selection of hrpY mutants. 24 25 Disruption of the fliC gene by plasmid insertion. A 0.4 kb SalI fragment including a 2 1 central part of the fliC gene was cloned from a PCR fragment amplified from RS1002 2 genomic DNA using primers P881 (5'-AGTCGCTGCAGCGCCTGTCGA-3') and P882 3 (5'-AAGGTCGACATGTTGACGTTG-3') into pARO191 to generate pRS845. pRS845 4 was integrated into the genome of R. solanacearum strains through conjugation with 5 S17-1. The transconjugants contain the pRS845 insertion into the chromosomal fliC 6 locus, resulting in mutants with disruptions of the fliC gene. 7 8 Construction of pilA mutants. A 0.5 kb fragment containing a 5'-half of the pilA gene 9 was PCR-amplified from RS1002 genomic DNA using primers 10 (5’-GGAATTCAAGTCGATGCCGGTCAAT-3’) 11 (5’-CGGGATCCGTCAGACTGTGCATTGGC-3’) with EcoRI (underlined) and BamHI 12 (double underlined) sites, respectively. Similarly, a 0.5 kb fragment containing a 3'-half 13 of 14 (5’-CGGGATCCGTCAGATCACCGTCACCT-3’) 15 (5’-AACTGCAGACGACCAGTTGCACGATC-3’) with BamHI (underlined) and PstI 16 (double underlined) sites, respectively. The two fragments were tandemly inserted into 17 pK18mobsacB, and a 2.9 kb BamHI Gmr gene cassette was inserted between them to 18 yield pRS844. Using pRS844, the pilA::Gmr mutation was introduced into R. 19 solanacearum strains by the marker-exchange method. The inactivation of pilA was 20 confirmed by the loss of twitching motility on plates (Kang et al., 2002). pilA was amplified and P814 using primers and P815 P818 P819 21 22 Construction of a popA-lacZYA fusion. Plasmid pINT102, which has the same 23 construction as that of pINT101 except for the opposite insertional direction, was used 24 to clone the prhA region as described above. The 2.3 kb SmaI fragment containing a part 25 of prhA was subcloned into pARO191. The resulting pINT103 was used to clone the 3 1 popA region from RS1002. The 3.3 kb SphI-BglII fragment including popA was 2 subcloned into pK18mobsacB to yield pRS188. Then, the promoterless lacZYA fragment 3 from pUC-lacZYA was inserted into the BamHI site in popA, yielding pRS192. 4 pUC-lacZYA was constructed as follows. The BamHI Kanr cassette was inserted into 5 pRS415 (Simons, 1987) to yield pRS415-BK. The promoterless lacZYA fragment was 6 excised using SalI from pRS415-BK and cloned into the multicloning site of pKRP10 to 7 yield pUC-lacZYA. RS1087 was constructed by introducing the popA-lacZYA fusion 8 into RS1085 by the marker-exchange method using pRS192. 9 10 Construction of hpx'-'lacZ fusions. To construct in-frame hpx'-'lacZ fusions, a 11 plasmid-based method was applied as follows. Approximately 1 kb restriction fragment 12 or PCR-ampliphied fragment containing a part of the coding sequence of hpx gene was 13 inserted into the SmaI site of plasmid pMC1871 to yield an in-frame hpx'-'lacZ fusion. 14 pMC1871 was contructed by inserting a 2.2 kb EcoRV-SmaI (Smr/Spr) cassette 15 from pHRP315 into the SmaI site of pMC1871 (Shapira et al., 1983). The resulting 16 plasmid carrying the hpx'-'lacZ fusion was integrated into the genome of R. 17 solanacearum strains through electroporation of the plasmid DNA (2 g). The 18 transformants contain the plasmid insertion into the chromosomal hpx locus, resulting in 19 hpx'-'lacZ fusions. 20 21 References 22 Parke, D. (1990) Construction of mobilizable vectors derived from plasmids RP4, 23 pUC18 and pUC19. Gene 93: 135-137. 24 Schäfer, A., Tauch, A., Jäger, W., Kalinovski, J., Thierbach, G., and Pühler, A. (1994) 25 Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli 4 1 plasmids pK18 and pK19: selection of defined deletions in the chromosome of 2 Corynebacterium glutamican. Gene 145: 69-73. 3 Shapira, S.K., Chou, J., Richard, F.V., and Casadavan, M.J. (1983) New versatile 4 plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ 5 gene sequences encoding an enzymatically active carvoxyl-terminal protein of 6 -galactosidase. Gene 25: 71-82. 7 5