SOP for detecting Ribosomal Slippages:
advertisement

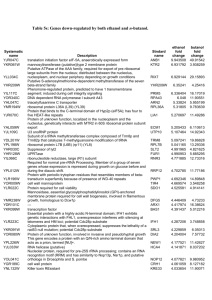
Standard Operating Procedure (SOP) for detecting Ribosomal Slippages Natalia Mikhailova, Genome Biology Program, JGI SOP for detecting ribosomal slippage for “peptide chain release factor 2” 1. Check if the gene annotated as “peptide chain release factor 2”, is shorter then some of the homologs. If it is, then very likely there is a ribosomal slippage in that gene. 2. Find the start sequence by performing blastx of the nucleotide sequence upstream of the gene. 3. To locate the exact place of the slippage try to find the nucleotide sequence Ctttgac The ribosomal slippage would be ctt-t-gac (it also could be ctt-t-aac) Ctt would be on the one frame, and gac – on the different frame. 4. Merge frames and determine a new start codon by the alignment of the homologous genes. 5. Annotate an extended gene in Artemis file as /exception="ribosomal slippage" References 1. Craigen,W.J., Cook,R.G., Tate,W.P. and Caskey,C.T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2 Proc. Natl. Acad. Sci. U.S.A. 82 (11), 3616-3620 (1985) 2. Lee,C.C., Kohara,Y., Akiyama,K., Smith,C.L., Craigen,W.J. and Caskey,C.T. Rapid and precise mapping of the Escherichia coli release factor genes by two physical approaches J. Bacteriol. 170 (10), 4537-4541 (1988) 3. Yanga Byuna, Sanghoon Moona and Kyungsook Han A general computational model for predicting ribosomal frameshifts in genome sequences Computers in Biology and Medicine Volume 37, Issue 12, December 2007, Pages 1796-1801 4. Michael Chandler and Olivier Fayet Translational frameshifting in the control of transposition in bacteria Molecular Microbiology Volume 7 Issue 4 Page 497-503, February 1993 SOP for detecting ribosomal slippages in transposases: 1. If a gene reported as “short” is annotated as a transposase, then check if the upstream gene is annotated also as a transposase. 2. If upstream gene is annotated also as a transposase, then blastx the nucleotide sequence of the region beginning at the start of the upstream gene and ending at the end of the short gene. 3. If the blastx gives long versions of the transposase, locate the place of the frameshift by looking at the pair-wise amino acid alignment. 4. Check if that place has a potential for a ribosomal slippage. That place should contain a heptanucleotide slippery sequence XXXYYYN (where X=A,G or T and Y=A or T) 5. If the location of the frameshift has a potential for a ribosomal slippage, then merge the genes and annotate the resulting gene as having a frameshift: /exception="ribosomal slippage" References 1. Yanga Byuna, Sanghoon Moona and Kyungsook Han A general computational model for predicting ribosomal frameshifts in genome sequences Computers in Biology and Medicine Volume 37, Issue 12, December 2007, Pages 1796-1801 2. Sanghoon Moon, Yanga Byun, Hong-Jin Kim, Sunjoo Jeong, and Kyungsook Han Predicting genes expressed via −1 and +1 frameshifts Nucleic Acids Res. 2004; 32(16): 4884–4892. Published online 2004 September 15. doi: 10.1093/nar/gkh829 3. Olga L. Gurvich, Pavel V. Baranov, Jiadong Zhou, Andrew W. Hammer, Raymond F. Gesteland and John F. Atkins Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli The EMBO Journal (2003) 22, 5941–5950 doi:10.1093/emboj/cdg561 4. Nina Mejlhede, John F. Atkins, and Jan Neuhard Ribosomal −1 Frameshifting during Decoding of Bacillus subtilis cdd Occurs at the Sequence CGA AAG Bacteriol. 1999 May; 181(9): 2930–2937 5. Michael Chandler and Olivier Fayet Translational frameshifting in the control of transposition in bacteria Molecular Microbiology Volume 7 Issue 4 Page 497-503, February 1993 A-AAA-AAG (From publication 5.) Examples of frameshift sequences CCCCCCTTTTT Transposase AAAAAAGGGGGG GGGGGAAAAAAA GGGGGAAACAAA TTTTTCCCCC (AAAAAGGGGG) TTGTTCCCCC (AACAAGGGGG) AAAAAGGGGG