Phenol Chloroform DNA Extraction
advertisement

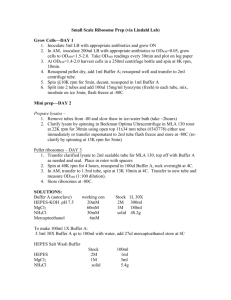
PROTOCOLS Phenol Chloroform DNA Extraction (Linton et al. 1999). Single mosquito are homogenised in a 1.5 eppendorf tube using a plastic pestle (Anachem), in 100 ml of extraction buffer containing 0.2 M sucrose, 0.1 M Tris– HCl, 50 mM EDTA and 0.5% SDS (Samson and Wagnez, 1980). Between successive extractions, plastic pestles are soaked in 0.25 mM HCL, to avoid cross-contamination of DNA samples and then washed thoroughly in distilled H2O, to get rid of any traces of HCL. The mixture is heated at 65°C for 10 minutes in a Techno Dry-Block® DB-3A (Genetic Research Instrumentation Ltd, England), 24 μl of 5mM KOAc (pH 9.0) is added and the tubes are placed on ice for a further 10 minutes. Following centrifugation at 13,200 rpm for 10 minutes, the supernatant is transferred to a clean 1.5 ml eppendorf, then 1 μl of 20 ug / ml RNAse is added and the tubes are transferred to a 37°C oven for 30 minutes. Following 100 μl of PCA (Phenol/Chloroform / Isoamyl, 25:24:1) is added and the tubes well vortexed, before centrifuging at 13.200 rpm for 1 minutes. The phenol facilitated the precipitation of fats and lipid membranes and the chloroform acted to break down proteins. The supernatant is removed, transferred to new eppendorf and 100 μl of chloroform added before again vortexing and centrifuging for 1 minute. This additional chloroform extraction ensured complete removal of phenol from the purified DNA, which could otherwise inhibit the Taq DNA polymerase in subsequent PCR reactions. Eighty μl of each supernatant is transferred to a new eppendorf and eighty μl of isopropanol is added, and samples stored at – 20 °C overnight. Samples are removed from the freezer and centrifuged at 13.200 rpm for 10 minutes. Isopropanol is decanted and the precipitate washed with 100 μl of cold 70% ethanol and spin for 10 minutes. The ethanol is decanted and the ethanol wash repeated with cold 100% ethanol. Ethanol is again removed, and the samples are desiccated in a speed vacuum for 20 minutes or in the over 37° for 30 minutes. 100 μl of 10mM Tris-HCl, 0.1 mM EDTA (Ph8.0) buffer (or dH2O) is added and the DNA left to resuspend at room temperature overnight. Following total resuspension, the tube are carefully labelled and stored in 200 μl PCR tubes (Greiner Labortechnik Ltd, Glos, England) at -70 °C until required. Isolation of total DNA from Animal Tissues (Qiagen). 1. Place the abdomen in a 1.5 ml microcentrifuge tube and add 180 μl Buffer ATL. 2. Add 20 μl proteinase K, mix by vortexing and incubate at 55°C for 3 hours. The samples can be lysed overnight; this will not affect them adversely. 3. Vortex for 15 seconds. Add 200 μl Buffer AL to the sample, mix thoroughly by vortexing and incubate at 70°C for 10 minutes. 4. Add 200 μl ethanol (96-100%) to the sample, and mix thoroughly by vortexing. 5. Pipet the mixture into the DNeasy mini spon Column placed in a 2 ml collection tube. Centrifuge at 8000 rpm for 1 min. Discard flow-trough and collection tube. 6. Place the DNeasy mini spin column in a new 2 ml collection tube, add 500 μl Buffer AW1, and centrifuge for 1 min at 8000 rpm. Discard flow-through and collection tube. 7. Place the DNeasy mini spin column in a 2 ml collection tube, add 500 μl buffer AW2, and centrifuge for 3 min at 14000 rpm to dry the DNeasy membrane. Discard the flow-through and collection tube. 8. Place the DNeasy mini spin column in a clean 1.5 ml or 2 ml microcentrifuge tube and pipet 200 μl buffer AE directly onto the DNeasy membrane. Incubate at room temperature for 1 min and then centrifuge for 1 min at 8000 rpm to elute. 9. Store the tubes until it will be used. You can to decide which to use (Phenol – Chloroform or Qiagen). Master mix for PCR reactions Mix for one reaction using Taq Polimerase (Bioline) 25.5 μl ddH20 5 μl 5.8 S primer a 5 uM 5 μl 28 S primer a 5 uM 5 μl dNTPs a 2 uM 5 μl 10xNH4 Buffer 2.5 μl 25mM MgCl2 0.1 μl Taq polymerase (BioLine) Vortex well and for each PCR adds 2 μl DNA plus 48 μl master mix. Note: You can to put one or two legs of the mosquito in the PCR mix directly without DNA extraction, it works very well (48 ul of master mix pluss one or two legs). THERMOCYCLER CONDITIONS ITS2 (5.8 S- 28 S) COI (Ubc 6 – Ubc 9) 1. 94°C for 2 minutes 2. 94 °C for 30 seconds 3. 57 °C for 1 minute 35 times 4. 72 °C for 30 seconds 6. 72 °C for 10 minutes 7. 10 °C hold Primers ITS2 (Collins & Paskewitz, 1996) 5.8 S: ATC ACT CGG CTC GTG GAT CG 28 S: GAC TAC CCC CTA AAT TTA AGC AT Sequence Reaction 1-2 ng DNA per 100 bps of product 1 pMol of Primer 3 μl Big Dye Dilution Buffer form Kit 1 μM big Dye Terminador Mix ddH2O up to 10 μl Cycle Sequence To do a hot start without the Big Die Terminator Reaction Mix for 5 minutes at 96 °C (step 1). The samples are then transferred to ice and the Big Dye added and mixed by pipetting up and down 2 or 3 times. The samples are returned to the Thermal Cycler for follow with the cycle sequences (step 2). 1. 96 °C for 5 minutes 2. 96 °C for 10 seconds 3. 50 °C for 5 seconds 25 Cycles 4. 60 °C for 4 minutes 5. Cool to 4 °C SOLUTIONS Grinding Buffer (Linton et al. 2001) To make 50 ml: 5 ml Tris-HCl at pH 8.0 3.42 g sucrose 2.5 g SDS 0.86g EDTA Fill to 50 ml with ddH20