Suggested sample preparation of gene expression array
advertisement

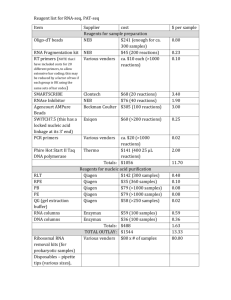
Gene expression 實驗 Suggest method and kit RNA isolation species or tissue Purification of total RNA (2nd) total RNA polyA-RNA Yeast Hot phenol protocol. (Schmitt, et al.,1990, Nucl Acids Res, 18:3091-3092.) Qiagen's Oligotex mRNA Kit Qiagen's RNeasy Mini Kit Arabidopsis TRIzol Reagent (Invitrogen Life Tech.) Qiagen's Oligotex mRNA Kit Qiagen's RNeasy Mini Kit Mammalian cells (cultured cells and lymphocytes) Qiagen's RNeasy Mini Kit Qiagen's Oligotex Direct mRNA Kit (directly from cell) or Qiagen's Oligotex mRNA Kit (from total RNA) N/A Mammalian tissue TRIzol Reagent from Invitrogen Life Tech. poly-A mRNA isolation commercial kit (from total RNA) Qiagen's RNeasy Mini Kit Albino or polysacchoride rich in plant Pine tree method N/A N/A (Adapted from GeneChip® Expression Analysis Technical Manual, Affymetrix company) RNA quality check by electron gel 1 2 3 4 5 For Electrophoresis: Please refer to the image at the left for the layout of the gel Lane 1:100 bp DNA ladder Lane 2: acceptable quality Lane 3: heavily degraded total RNA Lane 4: slightly degraded and DNA- contaminated sample Lane 5: 1 Kb DNA ladder Prokaryotic Total RNA Isolation As starting material for the cDNA synthesis procedure, total RNA can be isolated by using standard procedures for bacterial RNA isolation or various commercial RNA isolation kits. For Pseudomonas aeruginosa and E. coli, we have successfully used the QIAGEN® RNeasy Mini Purification Kit. Caution should be used to minimize chromosomal DNA contamination during the isolation, due to the high sensitivity of the assay. It is suggested that no more than 1 X 109 cells are applied to a single purification column. Also, use the lysozyme at a concentration of 1 mg/mL, and not the recommended 400 μg/mL. Additional DNase I treatment may be required to eliminate DNA contamination when the bacterial culture is grown at high density. After purification, RNA concentration is determined by absorbance at 260 nm on a spectrophotometer (1 absorbance unit = 40 μg/mL RNA). The A260/A280 ratio should be approximately 2.0, with ranges between 1.8 to 2.1 considered acceptable. We recommend checking the quality of RNA by running it on an agarose gel prior to starting the assay. The 23S and 16S rRNA bands should be clear without any obvious smears. Any indication of the presence of chromosomal DNA contamination (high molecular weight bands or smears on the gel) would require additional DNase treatment before proceeding to cDNA synthesis.