Name
advertisement

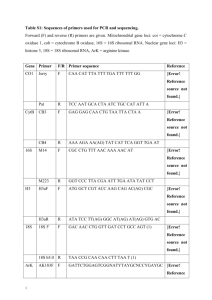
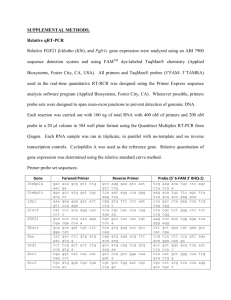
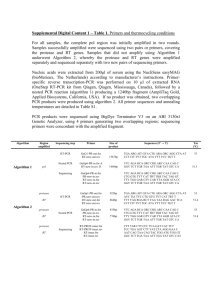
Bio102: Introduction to Cell Biology and Genetics Key for BioSynthesis on Decoding DNA Below is a 200 base-pair (bp) region of the chromosome of the bacterium E. coli. This region of the chromosome includes the gene kdpF, which encodes the shortest known protein in E. coli, a subunit of a membrane transporter. The sequence of each DNA strand is written three times to show that there are three possible reading frames (three ways to read potential codons) on each strand. For example, the sequence ATCCGA could be read ATC|CGA, A|TCC|GA, or AT|CCG|A. How can we identify a gene in a DNA sequence? One way is to look for open reading frames (ORFs): sequences of codons starting with a start codon and ending with a stop codon. An open reading frame of reasonable length could be a gene. In the sequence below, find all of the possible ORFs by circling each start codon, underlining the codons in the open reading frame, and putting a box around the stop codon at the end of the ORF. There can be more than one ORF in each of the six reading frames. 5′ 5′ 5′ 3′ 3′ 3′ TTG CAG CCA GAA TTC TAC CCT TCC GGT ATC ACT TTT AGG CCA CTG GAG GTG CAC TAT TT GCA GCC AGA ATT CTA CCC TTC CGG TAT CAC TTT TAG GCC ACT GGA GGT GCA CTA T T TGC AGC CAG AAT TCT ACC CTT CCG GTA TCA CTT TTA GGC CAC TGG AGG TGC ACT AT AAC GTC GGT CTT AAG ATG GGA AGG CCA TAG TGA AAA TCC GGT GAC CTC CAC GTG ATA AA CGT CGG TCT TAA GAT GGG AAG GCC ATA GTG AAA ATC CGG TGA CCT CCA CGT GAT A A ACG TCG GTC TTA AGA TGG GAA GGC CAT AGT GAA AAT CCG GTG ACC TCC ACG TGA TA 57 57 57 57 57 57 58 58 58 58 58 58 GAG TGC AGG CGT GAT AAC CGG CGT ATT GCT GGT GTT TTT ATT ACT GGG TTA TCT GGT GA GTG CAG GCG TGA TAA CCG GCG TAT TGC TGG TGT TTT TAT TAC TGG GTT ATC TGG T G AGT GCA GGC GTG ATA ACC GGC GTA TTG CTG GTG TTT TTA TTA CTG GGT TAT CTG GT CTC ACG TCC GCA CTA TTG GCC GCA TAA CGA CCA CAA AAA TAA TGA CCC AAT AGA CCA CT CAC GTC CGC ACT ATT GGC CGC ATA ACG ACC ACA AAA ATA ATG ACC CAA TAG ACC A C TCA CGT CCG CAC TAT TGG CCG CAT AAC GAC CAC AAA AAT AAT GAC CCA ATA GAC CA 114 114 114 114 114 114 115 115 115 115 115 115 TTA TGC CCT GAT CAA TGC GGA GGC GTT CTG ATG GCT GCG CAA GGG TTC TTA CTG ATC TT ATG CCC TGA TCA ATG CGG AGG CGT TCT GAT GGC TGC GCA AGG GTT CTT ACT GAT C T TAT GCC CTG ATC AAT GCG GAG GCG TTC TGA TGG CTG CGC AAG GGT TCT TAC TGA TC AAT ACG GGA CTA GTT ACG CCT CCG CAA GAC TAC CGA CGC GTT CCC AAG AAT GAC TAG AA TAC GGG ACT AGT TAC GCC TCC GCA AGA CTA CCG ACG CGT TCC CAA GAA TGA CTA G A ATA CGG GAC TAG TTA CGC CTC CGC AAG ACT ACC GAC GCG TTC CCA AGA ATG ACT AG 171 171 171 171 171 171 172 172 172 172 172 172 GCC ACG TTT TTA CTG GTG TTA ATG GTG C GC CAC GTT TTT ACT GGT GTT AAT GGT GC G CCA CGT TTT TAC TGG TGT TAA TGG TGC CGG TGC AAA AAT GAC CAC AAT TAC CAC G CG GTG CAA AAA TGA CCA CAA TTA CCA CG C GGT GCA AAA ATG ACC ACA ATT ACC ACG 3′ 3′ 3′ 5′ 5′ 5′ 1. Highlight the ORF that you think is most likely to be the kdpF gene with a highlighter pen, or circle it in a different color. Why did you choose this ORF? All of the possible ORFs are shown above, with start codons in dark blue and stop codons in red; the 5 different ORFs are highlighted in yellow, light blue, teal, green and gray. There are only two ORFs with both a start and stop codon within this sequence; the gray one is way too short to be a gene of any kind; the most likely choice is the one highlighted in yellow. 2. Name a DNA sequence that we could look for as additional evidence that one or more of these ORFs is actually a gene that can be translated in E. coli. There would be a promoter sequence somewhere upstream, but it could be fairly far away; the most immediate indication that it’s translated would be a Shine-Dalgarno sequence that the ribosome could bind just before the start codon. The standard Shine-Dalgarno sequence is AGGAGG, and there is a good match for this sequence just before the start of the yellow ORF (boxed in black). 3. If this were a region of eukaryotic DNA, you’d be very unlikely to find an actual gene gene by looking for an ORF. Why is this true? Introns disrupt coding sequences frequently in eukaryotes—almost every gene in higher eukaryotes. There could be stop codons within the intron which would fool us into thinking we’d found an ORF, when actually it continues on after splicing.