Sugar_Nucleotide_extraction_using_Methanol
advertisement

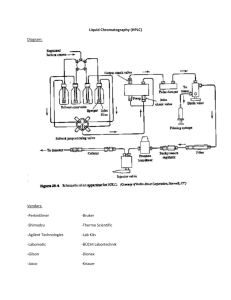
Sugar Nucleotide extraction using Methanol-Chloroform followed by quantitative Dionex-based HPLC analysis Partha Banerjee & Koen Raedschelders Introduction This protocol and its accompanying video are meant to provide an overview of the preparative and instrumental steps involved in the analysis of sugar nucleotides from cell extracts. The full process can be divided into three sequential phases: 1. Harvesting cells (not shown in the video) 2. Methanol-Chloroform based Sugar nucleotide extraction 3. Dionex-based HPLC analytical separation This protocol will cover the Methanol-Chloroform extraction phase. Quantitative sugar nucleotide analysis from cells provides useful information about fundamental biological processes that include glycan synthesis, metabolism, nucleotide flux, and in the case of O-GlcNAc, intracellular signal transduction. The Methanol-Chloroform extraction results in three distinct phases; a) Upper aqueous phase –containing nucleotide sugars and metabolites b) Interphase –containing the insoluble protein disc c) Lower organic phase –containing hydrophobic metabolites and lipids For the purposes of this protocol, the upper aqueous phase needs to be isolated and subjected to HPLC analysis. The insoluble protein disc can also be collected so that nucleotide sugars from a given can be normalized to their respective protein equivalents. Materials and equipment 1. Methanol:Water (2:0.8) On ice 2. MilliQ Water 3. Chloroform 4. Labelled 1.5 mL tubes 5. Sonicator 6. Centrifuge 7. 0.22um PVDF spin filter 8. HPLC analysis system (ie. Dionex 3000 LC system) Procedure Deriving pellets from cells 1. Wash cells with ice cold PBS 2. Gently scrape cells and transfer to labeled centrifuge tube 3. Pellet cells at 1000g for 10 minutes, remove supernatant, and store pellets at -80oC until required Methanol-Chloroform phase separation 1. Thaw pellets on ice briefly 2. Add 0.280ml of Methanol:Water to cell pellet and vortex 3. Sonicate for 5 seconds 4. Add 1 volume of chloroform, vortex 5. Add 1 volume of MilliQ, vortex 6. Centrifuge at 4oC for 5minutes, at 18,000xg 7. Transfer the upper aqueous layer containing sugar nucleotides to a new labeled tube i. Dry the aqueous layer (speedvac or lyophilize) ii. Resuspend in water containing internal standard (i.e. ~70M BrdU) iii. Filter through a 0.22 m PVDF spin filter iv. Analyze by HPLC 8. Transfer the protein to a new labeled tube i. Resuspend the protein in 50mM Tris, 0.5% SDS ii. Perform a protein estimation iii. Record these values and use them to normalize sugar nucleotide peaks 9. Save the lower organic layer if analysis of hydrophobic metabolites is desired Dionex-based HPLC separation of sugar nucleotides 1. Generate a suitable HPLC workflow (software dependent; refer to video for details) HPLC Solvents: i. Solvent A = 1M NaAc in 100mM NaOH ii. Solvent B = 100mM NaOH iii. Solvent C = HPLC-grade water -Gradients and separation conditions are column and system dependent. -Be sure to run a blank (BrdU only) to determine noise, and internal standard migration time.