file - BioMed Central
advertisement

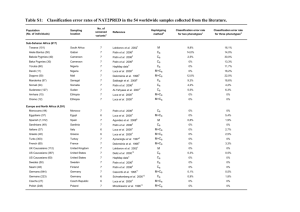
Additional file 1: Summary description of the 41 samples included in the worldwide genotyping survey Pop. a Code Sampling location Population (No. of individuals) Description No. of screened variants Nhb Hc sub-Saharan Africans (452) 1 Tswana (101)1 South Africa Tswana-speaking people from the North-West Province. They were all healthy volunteers and were part of Transition and Health during Urbanization of South Africans (THUSA) study. 7 12 0.8589 2 Ateke Bantus (50)2 Gabon Healthy donors 7 12 0.8091 3 Bakola Pygmies (40)2 Cameroon Healthy donors 7 7 0.7108 4 Baka Pygmies (30)2 Cameroon Healthy donors 7 8 0.8237 Nigeria Samples collected in a particular community in Ibadan, from individuals who identified themselves as having four Yoruba grandparents (HapMap sample). 6 7 0.8314 Senegal Healthy unrelated individuals from the Niohkolo Mandenka population. 7 11 0.8168 Mali Unrelated healthy non-caste Dogons collected in 6 villages in the district of Sangha, Republic of Mali. 7 12 0.8228 Somalia Healthy donors 7 6 0.7066 Morocco Healthy donors 7 6 0.6583 3 5 Yoruba (60) 6 Mandenka (97)4 5 7 Dogons (50) 8 2 Somali (24) d North Africans (44) 9 Moroccans (44)2 Europeans (3,570) 10 Spanish (258)6 Spain Healthy, unrelated white Spanish volunteers. All subjects were in good health and with no antecedent of disease. Most subjects were medical students from Extremadura (Badajoz, Spain) and the surrounding area. 7 8 0.6541 11 Sardinians (49)2 Sardinia Healthy donors 7 5 0.6230 France Unrelated, healthy French Caucasians. 7 7 0.7027 12 French (60) 5 13 French-Canadians (291) 14 UK Caucasians (112)1 15 US Caucasians (387) 8 16 17 7 e Canada Healthy controls randomly selected from a large institutional DNA bank. They were all of French origin and resided in the Province of Quebec, Canada. 5 7 0.6759 United Kingdom Healthy volunteers from the Cambridge area. 7 9 0.6883 United States Non-cancer women of European origin residents of Iowa, from a nested case-control study of the Iowa Women's Health Study. 7 9 0.7013 US Caucasians (60)3 United States Samples collected from people living in Utah with ancestry from northern and western Europe (HapMap sample). 6d 7 0.7265 Swedes (50)2 Sweden Healthy donors 7 6 0.6612 2 18 Saami (48) Finland Healthy donors 7 5 0.7149 19 Germans (844)9 Germany Unrelated subjects of German origin from Berlin. They include healthy volunteers (291) and hospitalized individuals (553) with various diseases but without known malignancy. 7 8 0.7208 20 Germans (223)10 Germany Healthy students and blood donors of the St. Jürgen Hospital (Bremen), from the northern part of Germany. 6f 8 0.7438 Additional file 1 (Continued) Pop. a Code Sampling location Population (No. of individuals) Description No. of screened variants Nhb Hc 21 Polish (248)11 Poland Unrelated children and adolescents of Polish origin. They were randomly selected from patients from the Wielkopolska region who had come to the Third Clinic of Children’s Diseases of the University School of Medicine in Poznan because of ordinary respiratory or urinary tract infections or during a routine control visit to the outpatient clinic. Patients with autoimmune disease or malignancy were excluded. 7 7 0.7461 22 Slovaks (167)12 Slovakia Healthy blood donors and patients over 65 years with rheumatological or cardiovascular diseases, at the Teaching Hospital Košice. They were all Slovak people from the general population of Eastern Slovakia. 5e 6 0.7386 23 Ashkenazi Jews (40)2 Healthy donors 7 4 0.6006 0.7157 24 Romanians (140) 25 Russians (290) 14 26 Turks (303)15 13 - f Romania Unrelated individuals from a Romanian population. 6 7 Russia Unrelated individuals from the European part of Russia (Voronezh region). Except for 71 healthy volunteers, subjects were outpatients of the Central Railway Clinic in Voronezh with non-malignant diseases. 7 7 0.7102 Turkey Unrelated Turkisk individuals from south-east Anatolia (born and living in Gaziantep and surrounding). Except for 18 healthy volunteers, all were outpatients of the Gaziantep University, Faculty of Medicine, with a broad range of non-malignant diseases. 7 7 0.7231 Central/South Asians (390) 27 Gujarati (50)2 India Healthy donors 7 8 0.7149 28 Turkmen (50) 2 Uzbekistan Healthy donors 7 6 0.7531 29 Kyrgyz (290)13 Kyrgyzstan Unrelated individuals living at Lake Issyk-Kul near China. 6f 8 0.7318 China Healthy Han Chinese from a farmer population in a remote rural area in Shanghai suburb. 7 6 0.5839 China Samples collected from individuals living in the residential community at Beijing Normal University who were self-identified as having at least three out of four Han Chinese grandparents (HapMap sample). 6 4 0.5735 East Asians (1,893) 30 Han Chinese (112)16 31 Han Chinese (45) 32 Chinese (44)2 3 d China Healthy donors from the Hunan and Zhejang regions. 7 5 0.6450 33 Japanese (144) 17 Japan Patients with rheumatoid arthritis randomly selected from the outpatient department of the Institute of Rheumatology, Tokyo Women’s Medical University, between 1992 and 1999. g 7 6 0.4376 34 Japanese (172)18 Japan Unrelated, ethnically Japanese women used as population controls. Those with non-Japanese ethnic parents were excluded. 6f 6 0.4823 Additional file 1 (Continued) Pop. a Code Sampling location Population (No. of individuals) Description No. of screened variants Nhb Hc 35 Japanese (44)3 Japan Samples collected in the Tokyo metropolitan area from people who came from (or whose ancestors came from) many different parts of Japan (HapMap sample). 6d 4 0.4846 36 Koreans (288)19 Korea Non-cancer women controls admitted to the department of surgery in three teaching hospitals located in Seoul from March 1994 to September 1999. 7 11 0.5599 37 Koreans (1,000)20 Korea Korean individuals who visited the health promotion center at Samsung Medical Center. 6f 11 0.5140 Thailand Healthy donors 7 5 0.7213 Panama Healthy volunteers, unrelated to the fourth degree of consanguinity, living in traditional Amerindian regions of Panama. Excluded from the samples were those subjects who did not speak an Amerindian language, presented any non-Amerindian physical characteristic, were not born in Amerindian regions, or whose parents did not meet these criteria. 6f 6 0.5672 Panama same description as Embera 6f 5 0.4229 Nicaragua Healthy, unrelated subjects from a mixed Nicaraguan population. Most of them were students and staff of the Universidad Nacional Autónoma de Nicaragua (León, Nicaragua). Only subjects with Central American Indianwhite mixed origin were included. All were in good health with had no history of serious disease. 7 11 0.7007 38 Thai (44) 2 Central Americans (378) 39 Embera (136)21 40 Ngawbe (105)21 41 a Nicaraguans (137) 22 Population code number Number of inferred haplotypes in the sample c Haplotype diversity d C282T missing e G191A and C282T missing f G191A missing g The frequencies of NAT2 haplotypes determined in this study on patients with rheumatoid arthritis were similar to those determined in Japanese controls b References 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. Loktionov A, Moore W, Spencer SP, Vorster H, Nell T, O'Neill IK, Bingham SA, Cummings JH (2002) Differences in N-acetylation genotypes between Caucasians and Black South Africans: implications for cancer prevention. Cancer Detect Prev 26:15-22. Patin E, Harmant C, Kidd KK, Kidd J, Froment A, Mehdi SQ, Sica L, Heyer E, Quintana-Murci L (2006) Sub-Saharan African coding sequence variation and haplotype diversity at the NAT2 gene. Hum Mutat 27:720. The International HapMap Consortium (2003) The International HapMap Project. Nature 426:789-796. This study Delomenie C, Sica L, Grant DM, Krishnamoorthy R, Dupret JM (1996) Genotyping of the polymorphic N-acetyltransferase (NAT2*) gene locus in two native African populations. Pharmacogenetics 6:177-185. Agundez JA, Olivera M, Ladero JM, Rodriguez-Lescure A, Ledesma MC, Diaz-Rubio M, Meyer UA, Benitez J (1996) Increased risk for hepatocellular carcinoma in NAT2-slow acetylators and CYP2D6-rapid metabolizers. Pharmacogenetics 6:501-512. Krajinovic M, Richer C, Sinnett H, Labuda D, Sinnett D (2000) Genetic polymorphisms of N-acetyltransferases 1 and 2 and gene-gene interaction in the susceptibility to childhood acute lymphoblastic leukemia. Cancer Epidemiol Biomarkers Prev 9:557-562. Deitz AC, Zheng W, Leff MA, Gross M, Wen WQ, Doll MA, Xiao GH, Folsom AR, Hein DW (2000) N-Acetyltransferase-2 genetic polymorphism, well-done meat intake, and breast cancer risk among postmenopausal women. Cancer Epidemiol Biomarkers Prev 9:905910. Cascorbi I, Drakoulis N, Brockmoller J, Maurer A, Sperling K, Roots I (1995) Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am J Hum Genet 57:581-592. Schnakenberg E, Lustig M, Breuer R, Werdin R, Hubotter R, Dreikorn K, Schloot W (2000) Gender-specific effects of NAT2 and GSTM1 in bladder cancer. Clin Genet 57:270-277. Mrozikiewicz PM, Cascorbi I, Brockmoller J, Roots I (1996) Determination and allelic allocation of seven nucleotide transitions within the arylamine N-acetyltransferase gene in the Polish population. Clin Pharmacol Ther 59:376-382. Habalova V, Salagovic J, Kalina I, Stubna J (2005) A pilot study testing the genetic polymorphism of N-acetyltransferase 2 as a risk factor in lung cancer. Neoplasma 52:364-368. Rabstein S, Unfried K, Ranft U, Illig T, Kolz M, Rihs HP, Mambetova C, Vlad M, Bruning T, Pesch B (2006) Variation of the Nacetyltransferase 2 gene in a Romanian and a Kyrgyz population. Cancer Epidemiol Biomarkers Prev 15:138-141. Gaikovitch EA, Cascorbi I, Mrozikiewicz PM, Brockmoller J, Frotschl R, Kopke K, Gerloff T, Chernov JN, Roots I (2003) Polymorphisms of drug-metabolizing enzymes CYP2C9, CYP2C19, CYP2D6, CYP1A1, NAT2 and of P-glycoprotein in a Russian population. Eur J Clin Pharmacol 59:303-312. Aynacioglu AS, Cascorbi I, Mrozikiewicz PM, Roots I (1997) Arylamine N-acetyltransferase (NAT2) genotypes in a Turkish population. Pharmacogenetics 7:327-331. 16. 17. 18. 19. 20. 21. 22. Guo WC, Lin GF, Zha YL, Lou KJ, Ma QW, Shen JH (2004) N-Acetyltransferase 2 gene polymorphism in a group of senile dementia patients in Shanghai suburb. Acta Pharmacol Sin 25:1112-1117. Tanaka E, Taniguchi A, Urano W, Nakajima H, Matsuda Y, Kitamura Y, Saito M, Yamanaka H, Saito T, Kamatani N (2002) Adverse effects of sulfasalazine in patients with rheumatoid arthritis are associated with diplotype configuration at the N-acetyltransferase 2 gene. J Rheumatol 29:2492-2499. Deguchi M, Yoshida S, Kennedy S, Ohara N, Motoyama S, Maruo T (2005) Lack of association between endometriosis and N-acetyl transferase 1 (NAT1) and 2 (NAT2) polymorphisms in a Japanese population. J Soc Gynecol Investig 12:208-213. Lee KM, Park SK, Kim SU, Doll MA, Yoo KY, Ahn SH, Noh DY, Hirvonen A, Hein DW, Kang D (2003) N-acetyltransferase (NAT1, NAT2) and glutathione S-transferase (GSTM1, GSTT1) polymorphisms in breast cancer. Cancer Lett 196:179-186. Lee SY, Lee KA, Ki CS, Kwon OJ, Kim HJ, Chung MP, Suh GY, Kim JW (2002) Complete sequencing of a genetic polymorphism in NAT2 in the Korean population. Clin Chem 48:775-777. Jorge-Nebert LF, Eichelbaum M, Griese EU, Inaba T, Arias TD (2002) Analysis of six SNPs of NAT2 in Ngawbe and Embera Amerindians of Panama and determination of the Embera acetylation phenotype using caffeine. Pharmacogenetics 12:39-48. Martinez C, Agundez JA, Olivera M, Llerena A, Ramirez R, Hernandez M, Benitez J (1998) Influence of genetic admixture on polymorphisms of drug-metabolizing enzymes: analyses of mutations on NAT2 and C gamma P2E1 genes in a mixed Hispanic population. Clin Pharmacol Ther 63:623-628.