Iron and the Pathogenicity of Bacteria
advertisement
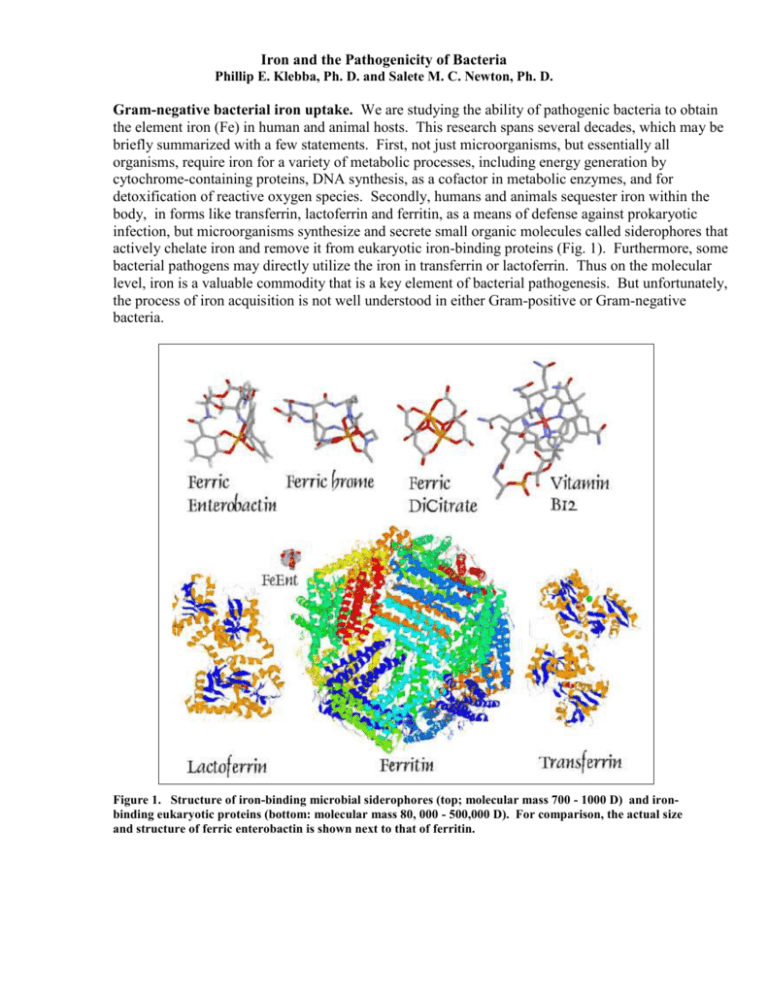
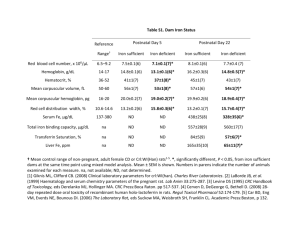
Iron and the Pathogenicity of Bacteria Phillip E. Klebba, Ph. D. and Salete M. C. Newton, Ph. D. Gram-negative bacterial iron uptake. We are studying the ability of pathogenic bacteria to obtain the element iron (Fe) in human and animal hosts. This research spans several decades, which may be briefly summarized with a few statements. First, not just microorganisms, but essentially all organisms, require iron for a variety of metabolic processes, including energy generation by cytochrome-containing proteins, DNA synthesis, as a cofactor in metabolic enzymes, and for detoxification of reactive oxygen species. Secondly, humans and animals sequester iron within the body, in forms like transferrin, lactoferrin and ferritin, as a means of defense against prokaryotic infection, but microorganisms synthesize and secrete small organic molecules called siderophores that actively chelate iron and remove it from eukaryotic iron-binding proteins (Fig. 1). Furthermore, some bacterial pathogens may directly utilize the iron in transferrin or lactoferrin. Thus on the molecular level, iron is a valuable commodity that is a key element of bacterial pathogenesis. But unfortunately, the process of iron acquisition is not well understood in either Gram-positive or Gram-negative bacteria. Figure 1. Structure of iron-binding microbial siderophores (top; molecular mass 700 - 1000 D) and ironbinding eukaryotic proteins (bottom: molecular mass 80, 000 - 500,000 D). For comparison, the actual size and structure of ferric enterobactin is shown next to that of ferritin. II. Gram-positive bacterial iron uptake. For all bacteria, the need for iron is problematical. Iron is unavailable in the aerobic microbial world, either because it is insoluble or because it is sequestered. In wild aqueous environments ferric iron rapidly precipitates as hydroxide polymers, and within animal hosts iron binding proteins like transferrin, lactoferrin and ferritin bind the metal with high affinity. Iron also circulates in the body as hemoglobin, which is normally ensconced within red blood cells. Indeed, iron is essential to the vast majority of organisms, but its sequestration by transferrin in serum and lymph, by lactoferrin in mucosal secretions, and by ferritin and heme compounds in cells, normally renders these fluids and tissues void of prokaryotic life. However, efficient pathogens overcome this barrier, by either producing siderophores or by utilizing iron-containing eukaryotic proteins. Many Gram-positive bacterial genomes were completely sequenced, including those of all the organisms listed above. All of the genomes contain loci with homology to iron transporters of Gram-negative bacteria (Fig. 2). These sequence data present a unique opportunity, the discovery of previously unknown and uncharacterized membrane transport proteins, with potentially different mechanisms than those of E. coli and its relatives. Figure 2. Putative iron transport loci in the genome of the Gram-positive organism L. monocytogenes. The indicated genes are fur regulated and show homology to ferric siderophore transporters of the E. coli inner membrane.