TPJ_4662_sm_figure-legends
advertisement

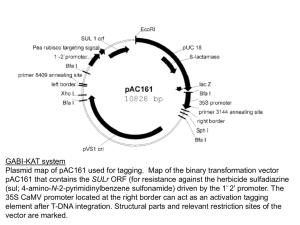
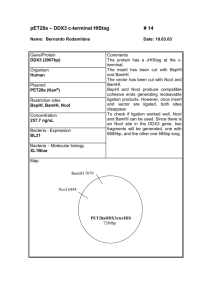
SUPPLEMENTARY FIGURE LEGENDS Figure S1: Assembly of binary vector constructs. (a) Intermediate vector pUBIAB-Cre; (b) control construct p6d35S-Cre5 (noTEL); (c) construct with one pAtT4-derived block of Arabidopsis-type telomeric repeats, p6d35S-Cre5-TEL7 (1xTEL); (d) construct with one pAtT4-derived block of Arabidopsis-type telomeric repeats, p6d35S-Cre5-2XTEL37 (2xTEL). Figure S2: Binary vector rearrangements. Rearrangement of constructs p6d35SCre5-TEL7 (1xTEL) and p6d35S-Cre5-2xTEL37 (2xTEL) during propagation in E. coli strain DH5. Primers used for sequencing analysis are indicated as arrows. Figure S3: Primer extension telomere repeat amplification (PETRA). (a) T-DNAassociated de novo telomere formation was detected by an adapted PETRA protocol. In step 1, PETRA-T oligonucleotides were annealed to the singlestranded G-rich overhang at the 3´end of the de novo telomeres and primer extension was performed with the help of DNA polymerase I. In the following step 2, the primer extension products were subjected to PCR amplification using the transgene-specific primer pUBI-F1 and the PETRA-T-complementary primer PETRA-A (Table S1) for a low number of cycles (20 times). The resulting PCRproducts were separated by gel electrophoresis, transferred to membrane and hybridised with transgene-specific probe (Step 3). (b) Confirmation of the specificity of the PETRA assay by Bal-31 exonucleolytic cleavage. Genomic DNA from plant T1#3-T2#11 known to contain internal T-DNAs and a terminal T-DNA associated with a telomere was incubated with Bal-31. No decrease of PCR product was observed for marker RB-At1g56600 specific for an internal T-DNA. In contrast, the PETRA signal for the terminal T-DNA co-segregating with marker At5g17440-LB decreased with increasing incubation with Bal-31. Thus, the PETRA assay specifically detects T-DNA-associated telomeres that are accessible to exonucleolytic cleavage. Figure S4: DNA gel blot confirmation of alternative modes of T-DNA-induced de novo formation of telomeres. (a) Schemes of different modes of T-DNA integration. A T-DNA with telomeric repeats at its right border (RB) can integrate as an internal T-DNA copy (top) or as a terminal T-DNA associated with a de novo formed telomere (middle, bottom). De novo telomere formation can initiate at different positions, either directly continuous with T-DNA-derived telomeric repeats (class I, middle) or leaving some T-DNA sequence between telomeric repeat blocks and RB in place (class II, bottom). The three different modes can be distinguished by DNA gel blot analysis using restriction enzymes NcoI and KpnI and hybridisation with a ProUBI-derived probe (orange bar). (b) Plant T1#3T2#6-T3#15 contains an internal T-DNA copy (Figure 3, Figure 6). NcoI cleavage does not result in a defined hybridisation signal. This is consistent with the presence of extended plant chromosomal DNA (not containing NcoI sites) continuous with the RB side of the T-DNA (Table S3). Combined NcoI and KpnI cleavage results in a defined hybridising fragment of approx. 2.0 kb length, confirming the presence of T-DNA sequence (including the KpnI site) from the region between the block of telomeric repeats and the RB. Plants T1#9-T2#14T3#S1 (S: grown on soil without selection) and T1#4-T2#2-T3#1 contain class I terminal T-DNA copies associated with newly formed telomeres (Figure 3, Figure 6). NcoI cleavage result in diffuse hybridisation signals (black vertical bars), indicating the presence of T-DNA-associated telomeres of approx. 5 kb and 6 kb length, respectively. Incubation with NcoI and KpnI cleavage does not alter signals, confirming the loss of the T-DNA region containing the KpnI site close to the RB in the course of telomere formation. Plant T1#3-T2#3-T3#13 contains a class II terminal T-DNA copy (Figure 3, Figure 6). NcoI cleavage results in a diffuse hybridisation signal, indicating the presence of a T-DNA-associated telomere of approx. 3 kb length. Combined cleavage with NcoI and KpnI results in a defined hybridising fragment of approx. 2.0 kb length, confirming the lasting presence of the T-DNA region close to the RB (including the KpnI site) between the T-DNA-derived telomeric repeat block and the newly added telomeric repeats.