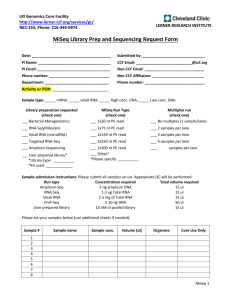
Sample Submission Form
advertisement

Illumina MiSeq Sample Submission Form Project number (filled by FMSC) Project title (with one sentence) Project Description CONTACT INFORMATION Contact person(s) Name(s) Email(s) Address Phone Group leader/Principal investigator Name Email Address Phone Biocity Turku Research Programme (must be filled by Biocity Turku groups) INVOICING INFORMATION Internal invoice (University of Turku account) Name Department/Laboratory Cost pool (kustannuspaikka) TY project code: 26xxxxx External invoice (other than University of Turku account) Institution Department/Laboratory Address Cost pool (kustannuspaikka), if applicable VAT number PROJECT OPENING MEETING Date Participants (FMSC + customer) MiSeq Sample Submission Form 2014 Page 1 of 7 MiSeq PROTOCOL INFORMATION Read length Analysis application type Number of lanes x 50 bp x 75 bp x 150 bp x 250 bp x 300 bp Targeted RNA-seq Targeted DNA-seq Custom Amplicon -seq Metagenomics (16S-seq) Small genome re-sequencing smallRNA-seq Clone-seq Other: ______________________ No. of runs used in sequencing: No. of samples per run: Additional library preparation protocol information (e.g. special kit) SAMPLE INFORMATION Total number of samples (including biological replicates) Organism Sample type Tissue, origin Cells, origin Blood, origin Other, what Sample material gDNA totalRNA poly(A) RNA smallRNA PCR Amplicon ready MiSeq library Other:_______________________ DNA / RNA extraction method Other possible pre-processing done to the samples (please specify all applied protocols) MiSeq Sample Submission Form 2014 Page 2 of 7 SAMPLE LIST Avoid spaces and any special characters in the sample names. Please use group names and sample names that will allow an easy interpretation of the name coding during data analysis. (Add rows if needed.) Also check the prices with FMSC if your total sample number has changed. Sample Sample name Sample group c, ng/l A260/A280 Volume, l Comments Index Pooling group 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 GROUP DESCRIPTIONS Please give a short description of each sample group (add rows if needed). Please note that if you have changed your original sample setup from the one discussed with our bioinformaticians, you should contact them (bioinfo@btk.fi) to discuss the changes before continuing further. This concerns especially those projects where the data analysis service is used. Sample group Num. of biological replicates Description COMPARISON TO A SET OF PREVIOUS OR FUTURE RESULTS If you are planning to compare these results with a previous set of results produced using our service, please indicate here the time when the previous samples were processed If you are planning to compare these results directly with a possible future set of samples that will be processed using our service, please indicate this here and also give us some estimate when these samples will be processed. (It should be noted that after longer time the same equipment and protocols may not be available anymore, complicating the integration between earlier and later data set) MiSeq Sample Submission Form 2014 Page 3 of 7 DATA ANALYSIS SERVICE I would like to purchase the full bioinformatics data analysis service (please contact the bioinformatics team for pricing and details bioinfo@btk.fi) Data will be analyzed through collaboration with the FMSC bioinformatics group (as agreed previously) I will analyze the data myself, data will be sent to this e-mail address: FILL IN DIRECT COMPARISONS BETWEEN GROUPS if FMSC's bioinformatics team will analyze the data Use group names given in the sample information table. Remember that all groups do not necessarily need to be directly compared but the lists of differentially expressed genes between several independent comparisons (e.g. for different tissues) can also be compared. Comparison Comparison group 0 (example) KO (knock-out group) Baseline group WT (wild type group) 1 2 3 4 5 6 7 8 9 10 Paired samples Please indicate clearly (by e.g. sample naming) the samples that originate from the same biological individual as this information is needed in the analysis. Other information for the bioinformatic analysis (e.g. other factors linking certain samples together like handling day etc.) MiSeq Sample Submission Form 2014 Page 4 of 7 COLLABORATION WITH FMSC This page is to be filled by only those with whom collaboration has previously been agreed. Type of collaboration Lab Bioinformatics Financing of the project (What does each group contribute to material and personnel, given as detailed as possible, if relevant include payment schedule) FMSC members to be included as coauthors to publication(s) Other issues (e.g. Secrecy, place for storage of samples, time plan for discarding remains of samples, use of remaining samples for other purposes than this project.) MiSeq Sample Submission Form 2014 Page 5 of 7 Sample submission requirements and service work agreement The sample requirements and sample preparation instructions are described below for the different types of samples. FMSC will always check the quality of the samples (or libraries) prior to sequencing and the customer is contacted if there is a concern related to sample quality. Please note that samples can be processed with customers consent even in the case they do not meet the quality requirements but in these cases FMCS will not be able to guarantee successful sequencing results. Please do not hesitate to contact ngs@btk.fi for any further questions. Ready-made libraries for sequencing All library concentrations and fragment profiles should be similar in the same sample set. If several libraries are pooled in one run, it is important to maintain color balance for each base of the index read for optimal de-multiplexing. FMSC personnel can assist in the selection of color balanced indexes before library preparation. Individual libraries prepared by the customer for pooling and MiSeq sequencing should be delivered in at least 4nM concentration and 10 µl volume. If the customer wants to pool his/her individual libraries themselves, the results of the concentration and size distribution measurement as well pipetting charts for pooling should be provided. Pooled and normalized libraries prepared with Nextera XT or TruSeq Custom Amplicon kits should be provided with a minimum volume of 25µl. RNA samples for library preparation and sequencing Samples are recommended to be delivered as total RNA in order to ensure purity and quality of the starting material before library preparation. It is recommended that only the required amount of RNA is sent since FMSC does not have extra storage space reserved for sample storage. All samples in the same set should be of equal purity and quality. The absorbance ratio A260/A280 for pure RNA is 2.0-2.2 and the integrity number(RIN) for intact total RNA above 8.0 Total RNA should be provided in 200 ng/µl and minimum volume of 10 µl. RNA that has DNA contamination will result in underestimation of the amount of RNA used. To prevent this, DNase step is recommended, but not required being included with the RNA isolation method. If RNA material meeting the criteria can’t be provided due to special circumstances such availability source material for RNA extraction alternative workflows for these cases exist and the possibilities should be discussed separately. DNA samples for library preparation and sequencing Samples are recommended to be delivered as genomic DNA in order to ensure purity and quality of the starting material before library preparation. It is recommended that only the required amount of DNA is sent since FMSC does not have extra storage space reserved for sample storage. All samples in the same set should be of equal purity and high quality. The absorbance ratio A260/A280 for pure DNA is 1.8-2.0. Genomic DNA samples should be diluted to 200 ng/ul concentration and in minimum volume of 10µl. DNA should be quantified using fluorescence based method such as Picogreen or Qubit. RNA contamination results in underestimation of the amount of DNA used. Therefore RNase step is recommended but not required to be included with the DNA isolation method. If PCR products are provided the estimated length of the products should be provided. If DNA material meeting the criteria can’t be provided due to special circumstances such availability source material for RNA extraction alternative workflows for these cases exist and the possibilities should be discussed separately. MiSeq sequencing run and result delivery The typical yield for a MiSeq run is 20-50 x 106 paired-end reads, depending on the version of sequencing reagents used. Sequencing will be rerun without extra costs in case of technical failure during the sequencing if it is not related to the nature or quality of the libraries. The raw data in Illumina’s default format (fastq) will be downloadable within a week after the sequencing has been finished. Sample and data storage FMSC will discard the left over RNA/DNA sample material after 1 year. Please deliver only the amount of sample needed for the analysis. If extra material is for some reason sent and customer wishes to have it returned, it has to be agreed separately. Customer has to cover the possible costs related to the returning of the sample material. FMSC will store data only for a short time and the customer is responsible for storing the raw data after the project is finished and the data has been delivered to the customer. MiSeq Sample Submission Form 2014 Page 6 of 7 Send us this sample submission form both in print with your samples and electronically as an e-mail attachment to ngs@btk.fi. We require a signed copy of this document prior to processing the samples. Please check detailed sample submission instructions on our website at http://fmsc.btk.fi. As Principal Investigator on this project, I agree to: 1. Acknowledge the use of the Finnish Microarray and Sequencing Centre's analysis service in the materials and methods section in any resulting publications. 2. Send a reprint of each resulting publication to the Finnish Microarray and Sequencing Centre. 3. Include all involved FMSC members (see previous page) to my resulting publication(s) (where the analysis results are used) as coauthors if collaboration has been agreed with them. I will also send the manuscript for their review prior to submitting to a journal. _______________________ Date MiSeq Sample Submission Form 2014 _______________________ Signature Page 7 of 7