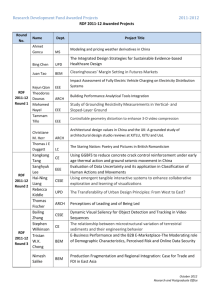
Supplementary Text (doc 3923K)
advertisement

“DNA copy number abnormalities do not occur in Infant ALL with t(4;11)/MLL-AF4” SUPPLEMENTARY TEXT CN and LOH analysis (Supplementary Figure 1 and 2). LOH analysis was performed by dChip software on Infant ALL diagnostic samples (n=28) paired with their corresponding remission (n=18) and relapse (n=8), when available. The homozygous regions were detected according to the following method: a matrix of single SNP probability of LOH was exported from dChip: an LOH event was assumed when the probability value was greater or equal to 0.5 (P≥0.5); we determined the stretches of consecutive, adjacent aberrant SNPs and then assigned the maximum values of LOH probability at these regions. To distinguish deletions from UPD we assessed the copy number of homozygous regions by combining LOH and CN data in genome view. For CN analysis with dChip software, the "mode copy number" option was applied, as a “cytonormalization" method to scale the inferred copy numbers by a factor of (2 / mode copy). Supplementary Fig. 1 shows a representative example of genomic profiling on chromosome 7 in which both UPD and deletions were observed in different patients. In the relapse sample of patient 9 and 21 the LOH associated to del(7p) was difficult to detect by dChip because of the presence of cellular heterogeneity. However, after visually inspection and CNAG analysis, the deletion was assigned although not accompanied by typical LOH signature (see also Fig. 2). The plot on Supplementary Fig.2 displays a selection of chromosomal regions (98, containing 15054 SNP markers) in which at least one event of UPD (in blue) occur among the diagnoses. For the vast majority of the cases for which the remission sample was available, the segmental UPD were constitutional, as detected at both diagnostic and remission phases. One patient (pt.9) displayed an exceptional high number of large UPD regions in both diagnostic and remission samples, which could likely be explained by familial consanguinity (not further investigated in this study). 1 Confirmation of segmental UPD regions by sequencing UPD(14)(q21.2) (Supplementary Figure 3). The UPD(14)(q21.2) is the most common UPD found in four out of twenty-eight patients: two patients (pt.8 and 21) were detected with both dChip and CNAG, two patients (pt.3 and 4) only with CNAG. The common altered region found in all the four patients is a 2,0 Mb segment (start: 44,213,211 end: 45,867,706) containing 3 genes, among them, the Fanconi Anemia complementation group M (FANCM) gene, involved in the DNA repair mechanism (30). The GTYPE visualization of raw genotype calls within the region under investigation clearly show the lack of heterozygous AB call (in green) in the 4 investigated Infant ALL patients (DX, REM and REL samples) compared to 6 healthy newborn used as normal controls (CNTRL1-6) (panel A). For all patients the stretch of homozygosity is detected at both diagnosis and remission level (constitutional), except for pt.21 (tumor associated), and persistent at relapse (in the two cases for which the sample was available) (panel B). The UPD(14)(q21.2) was detected in all cases by CNAG analysis (and shown in panel C as a representative example). Ultimately, the homozygous status of infant ALL diagnostic and remission samples compared to heterozygous normal controls was validated by direct sequencing of highly heterozygous SNP within the region under investigation (panel D). The rs226984 is a non-coding SNP mapping on FANCM gene locus. In only one case (pt.21 remission) the sequence results was found in disagreement with the SNP call (C.I.= 0.000488): according to the sequence the DNA of pt.21 at remission carries the homozygous A/A status identical to the correspondent diagnostic sample. The heterozygous AB call is probably a miscalling error of Affymetrix SNP genotyping array. Similarly, the heterozygous status of additional 18 coding (synonymous or nonsynonymous) SNPs within the coding sequence of FANCM gene (rs61746895, rs10138997, rs45547534, rs61753893, rs61745871, rs61744648, rs1367580, rs8017226, rs45568842, rs61748921, rs61749475, rs45604036, rs61730251, 2 rs61746943, rs7142192, rs3736772, rs45557033, rs8018014) was confirmed by direct sequencing the FANCM mRNA on the four patients with UPD(14)(q21.2) (data not shown). UPD(7)(q31.33q32.1) (Supplementary Figure 4). UPD(7)(q31.33q32.1) was detected in three out of twenty eight patients (pt.1,9 and 11) by dChip and CNAG analysis (see Supplementary Fig.3). Although the region is different in size among the three patients (much larger in pt.9), the minimal common altered segment is 1,7 Mb (start:125,763,796 end:127,562,585). In all the cases the UPD is constitutional (found at both diagnosis and remission level) and persistent at relapse in patiens for whom the samples were available for analysis (panel A, B, C). Four SNPs with high heterozygous frequency were selected within the region and sequenced to validate the homozygous status of the three patients under investigation compared to normal controls (panel D). UPD(8)(q21.13) and UPD(8)(q24.11) (Supplementary Figure 5 and 6). Chromosome 8q resulted to be the one mainly affected by UPD at high frequency. UPD(8)(q21.13) and (8)(q24.11), of 1,5Mb and 1,2Mb respectively, are recurrent in two out of twenty-eight patients at diagnosis each. The minimal common altered region on chr(8)(q21.13) (start: 78,089,129 end: 79,626,121) contains genes encoding for the protein kinase inhibitor protein specific for the catalytic subunit of cAMP-dependent (PKIA). On chr(8)(q24.11) (start: 117,807,670 end: 119,004,046) the nuclear matrix protein (RAD21), the extosin1 (EXT1) gene and the TRAP/mediator complex component TRAP25 (THRAP6) genes are mapping. The latter was found to be altered (del(8)(q24.11)) in one additional case (pt.22) (see Tab.2). The size of the lesion is similar among patients for UPD(8)(q21.13), different for (8)(q24.11) (larger in pt.9, smaller in pt.2). Whereas the remission/relapse samples were available for analysis the lesion was shown to be constitutional and persistent at relapse. (panel A, B). The chromosomal segments were confirmed by CNAG (panel C) and validated by direct sequencing (panel D). 3