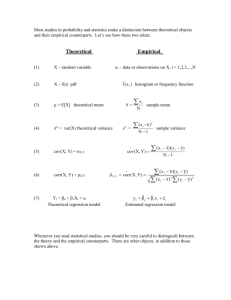
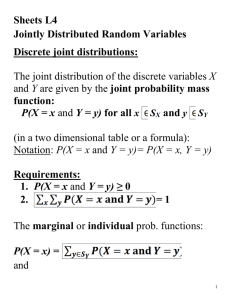
Genetics 422/515 Exercises on BLUP
advertisement

Exercises Day 1 Part 1: EBV accuracy Exercise 1 Effect of using relatives’ information on selection accuracy For single trait prediction of breeding value, write out the P-Matrix and the G-vector for the following cases: 1. One own performance record (X1) P-matrix: var(X1) = 2P is the phenotypic variance G-vector: Cov(X1, A) = Cov(A+E, A) = Cov(A, A) + cov(E,A) = 2A + 0 = 2A b = P-1G = (2P )-1 2A = h2 accurcay2 = b’G/2A = h2. 2A/ 2A = h2. Hence, accuracy = equal to h. 2. Information known on own performance and performance of sire (1 record each) Information sources: X1 = own performance X2 = performance of sire variance and covariance of information sources: cov( X1 , X 2 ) X var( X1 ) var 1 P var( X 2 ) X2 cov( X 2 , X1 ) var(X1) = 2P is the phenotypic variance var(X2) = 2P is the phenotypic variance Cov(X1,X2) = Cov(A+E, As +Es) = Cov(A, As) + cov(A,Es) + cov(E,As) +cov(E,Es) = ½2A+ 0 + 0 + 0. covariance between information sources and the animal’s breeding value X cov( X1 , A) cov( 1 , A G X2 cov( X 2 , A) Cov(X1, A) = 2A Cov(X2, A) = Cov(As +Es , A) = Cov(As, A) + cov(Es,A) = ½2A + 0 . such that index weights obtained by regression = covariance/variance: Exercise Day1 1 P2 b1 1 P G 1 2 b2 2 A 1 A2 A2 1 P2 1 2 A2 1 2 h2 1 2 1 1 h2 h2 1 1 2 h2 2 3. Information known on own performance and an EBV of the sire (accuracy = 0.9) variance and covariance of information sources: cov( X1 , X 2 ) X var( X1 ) var 1 P var( X 2 ) X2 cov( X 2 , X1 ) var(X1) = 2P is the phenotypic variance var(X2) = r22A where r is accuracy and r2 is reliability Cov(X1,X2) = ½ r2. 2A covariance between information sources and the animal’s breeding value X cov( X1 , A) cov( 1 , A G X2 cov( X 2 , A) Cov(X1, A) = 2A Cov(X2, A) = ½ r2 2A 4. Information known on own performance and an EBV of the sire (acc=0.9) and dam (acc=0.5) P is a 3 by 3 matrix, G a 3 by 1 vector 5. Information on own performance and 25 half sibs Information sources: X1 = own performance X2 = mean performance of n half sibs variance and covariance of information sources: cov( X1 , X 2 ) X var( X1 ) var 1 P X cov( X , X ) var( X 2 ) 2 1 2 var(X1) = 2P is the phenotypic variance var(X2) = tHS 2P + ((1-rHS)/n) 2P where tHS = 1/4 h2 Cov(X1,X2) = tHS is the intra class correlation covariance between information sources and the animal’s breeding value X cov( X1 , A) cov( 1 , A G X2 cov( X 2 , A) Cov(X1, A) = 2A Exercise Day1 2 Cov(X2, A) = ½ 2A such that index weights obtained by regression = covariance/variance: 1 1 P2 A2 1 r r P2 b1 h2 1 P G 2 2 1 2 2 b2 r P [r (1 r ) / n] P 2 A r [r (1 r ) / n] 1 2 h 6. Information on own performance, EBV of the sire (acc=0.9) and 25 half sibs 7. Information on own performance, 25 half sibs and 50 progeny Use the symbols VA for additive genetic variance and VP for phenotypic variance. Note that for a single trait prediction you can also substitute these by V P = 1 and VA = h2 You can plug the formulas from above in Excel, and for these cases calculate the index weights and the accuracy, assuming a certain value for h 2.. For some guidance on matrix calculation in Excel, see the end of this exercise sheet. You can check your answers with STSELIND.XLS The code looks like vrep = rep + (1 - rep) / nrep vdam = rep + (1 - rep) / ndam vsir = rep + (1 - rep) / nsir vfs = tfs + (1 - tfs) / nfs fsgrsize = nfs + 1 fsOD = (nfsgr - 1) * (fsgrsize * (fsgrsize - 1)) hsOD = (nhs * (nhs - 1)) - fsOD vhs = (nhs + fsOD * tfs + hsOD * ths) / (nhs * nhs) vprog = ths + (1 - ths) / nprog P(1, 1) = vrep P(1, 2) = 0.5 * h2 P(1, 3) = 0.5 * h2 P(1, 4) = tfs P(1, 5) = 0.25 * h2 P(1, 6) = 0.5 * h2 P(2, 2) = vdam P(2, 3) = 0 P(2, 4) = 0.5 * h2 P(2, 5) = 0 P(2, 6) = 0.25 * h2 P(3, 3) = vsir Exercise Day1 3 P(3, 4) = 0.5 * h2 P(3, 5) = 0.5 * h2 P(3, 6) = 0.25 * h2 P(4, 4) = vfs P(4, 5) = 0.25 * h2 P(4, 6) = 0.25 * h2 P(5, 5) = vhs P(5, 6) = 0.125 * h2 P(6, 6) = vprog For i = 2 To 6 For j = 1 To i P(i, j) = P(j, i) RHS(i, j) = RHS(j, i) RFS(i, j) = RFS(j, i) Next Next G(1, 1) = h2 G(2, 1) = 0.5 * h2 G(3, 1) = 0.5 * h2 G(4, 1) = 0.5 * h2 G(5, 1) = 0.25 * h2 G(6, 1) = 0.5 * h2 Exercise Day1 4 Exercise 2 Correlations between relatives’ EBV Consider the following cases, and for each case, calculate the correlation between EBVs on full sibs and half sibs. You can use STSELIND, and use the P-matrix and the index weights to work out this problem. 1. Information known on EBV of the sire (acc=0.9) and dam (acc=0.5) 2. One own performance record 3. Information known on own performance and an EBV of the sire (acc=0.9) and dam (acc=0.5) 4. Information on own performance, EBV of the sire (acc=0.9), dam (acc=0.5) and 50 progeny See the new stselind: ' R matrices for correlation among siblings RFS = P RHS = P RFS(1, 1) = tfs RFS(6, 6) = h2 / 8 RHS(1, 1) = ths RHS(1, 2) = 0 RHS(1, 4) = ths RHS(2, 2) = 0 RHS(2, 6) = 0 RHS(4, 4) = ths RHS(6, 6) = h2 / 16 Exercise Day1 5 Exercise 3 Pseudo BLUP In real life, parents have not just their own records, but they have an estimated breeding value with a certain accuracy, using BLUP. This accuracy is based on ancestor information, their own siblings and perhaps their offspring. The STSELIND program does not take these into account, other than that the progeny of the parents are the sibs of the animals. Also, BLUP corrects for the records of the mates of sires, when their progeny are evaluated. The amount of ancestral information can be derived from a given population structure. The program BLUPEBV.XLS does a full Pseudo-BLUP prediction of EBV accuracy, given genetic parameters, and a certain population structure (Half-sib and full-sib family size). In the following, use BLUPEBV to calculate EBV accuracy, and compare that with the more simple STSELIND approach: h2=0.25, FS family size =2, HS family =40 and h2=0.10 FS family size =4, HS family =80 Explain the negative weight on EBV of mates Vary h2 and look at weights on parental EBV Exercise Day1 6 Exercises Day 1 Part 2: Selection response Exercise 1 Consider the following breeding program that is being initiated in a large breeding herd: Selection is based on EBV derived from own performance only, for a repeated measures trait that is evaluated on both males and females at 6 months of age and at 18 months of age. Trait parameters are: h2=.30 repeatability = 0.6 Phenotypic st. dev. = 10 Each year 500 males and 500 females are born and available for selection at 6 months of age. Males stay in the herd for 1 year and are used for breeding for one breeding season (year). Each year, the top 50 6-month-old sires are selected based on EBV derived from own performance at 6 months of age. They produce their progeny when they are 1 year of age Females stay in the herd for 2 years but only 75% of all 1-year old females survive to 18 months of age; 25% of all 1-year-old females are culled from the herd for reasons unrelated to the trait under selection. So each year 500 6-month olds and 375 18-month-olds are available. Each year the top 125 out of the 500 6-month-old females and the top 75 out of the 375 18month-old females are selected for breeding. 6- and 18-month-olds are selected based on EBV from own performance at 6 months and on the average performance at 6- and 18-months, respectively. They produce progeny when they are 1 and 2 years of age, respectively. In year 0, the herd is unselected and consists of 500 6-month-old males, 500 6-month-old females, and 375 18- month-old females (NOTE: the herd is unselected, thus all age groups have the same genetic mean in year 0) Our aim is to predict the genetic progress that will be made in this herd over the next 2 years. a) Predict the average genetic level of progeny that are born in year 1 (i.e. based on selection of the 6-month-old males and the 6-month-old and the 18-month-old females that are present in year zero). Hint: predict the average genetic level of the selected 6-month-old sires, the selected 6-month-old dams, and the selected 18-month-old dams, and based on those, predict the genetic level of the progeny. b) Predict the average genetic level of progeny that are produced in year 2 (i.e. based on selection of sires that are 6 months old in year 1 and females that are 6 and 18 months old in year 1). Hint: use the same procedure as under a) but now take into account that the males (females) that are now 6-months old, are the progeny whose mean you calculated under a). c) Predict the asymptotic rate of progress per year for the breeding program described above and compare it to the progress that is made from year to year based on the calculations made in a) and b). Discuss your results. In year 1, the breeder now decides to optimize the proportions selected from 6- and 18-monthold females, for a total of 200 dams selected, rather than using 125 6-month-olds and 75 18month-olds, in order to maximize the genetic level of progeny in year 2. d) Using TRUNCSEL.XLS, determine the optimal proportions selected from the two age groups of females that are available in year 1. To do this, specify the mean and standard deviations of the EBV and the proportion of available selection candidates (females) that are 6- and 18months old, and enter this in the spreadsheet. e) Using the proportions determined in d), compute the genetic level of progeny born in year 3 and compare this with results obtained in c). Exercise Day1 7 Exercise 2 The response per year can be given by the formula of Rendel and Robertson: Ryr = S/L. For simplicity assume equal selection intensities in males and females). We can maximize selection response by truncation section across age classes, (assuming the selection criteria are comparable across age classes). Truncation selection across age classes maximizes the mean of the selected parents. Show algebraically that maximizing the mean of selected parents results in maximizing the response per year (hence, optimizes selection across age classes) Exercise 3 Using the Excel Spreadsheet ‘Genetic_gain.xls’, evaluate the impact of the percentage of cows inseminated by young bulls and progeny group size on genetic gain. Find the optimal combination of these two variables in order to maximize genetic gain. Exercise Day1 8 Exercises Day 1 Part 3: Change of Variance Exercise 1 Consider a trait with heritability equal to 0.25 and phenotypic standard deviation equal to 20. The trait is expressed in females only. Assume the infinitesimal genetic model. In a population of infinite size, each generation the best 10% males and the best 50% females are selected and mated at random to produce the next generation. Males are selected based on the average performance of 5 half-sisters. Females are selected on an optimal index of own performance and the average performance of 4 half-sisters. There are no common environmental effects among sibs. All half sisters are themselves half sibs of each other. a. Starting from an unselected population, derive accuracies of selection and response to selection in the initial generation (=generation 1). b. Taking into account the Bulmer effect, derive the genetic variance among progeny that result from selection in that first generation. c. Taking into account the effect of selection on genetic variance and accuracy of selection due to the Bulmer effect, predict the accuracies of selection (i.e. set up the appropriate indexes based on the new genetic variances), responses to selection, and genetic variance of progeny that result from selection in the second generation. Discuss the results. d. Repeat b. and c. but now assume that selection is on Animal Model BLUP EBV. The accuracies of EBV used for selection are 0.25 for males and 0.529 for females. Note: these accuracies are the accuracies that are obtained from the inverse of the Mixed Model Equations without accounting for the effect of selection on genetic variance (i.e. they are accuracies based on an unselected population). e. Derive the asymptotic genetic variance, accuracy, heritability and response to selection for the breeding program under d, using Animal Model BLUP EBV. f. Compare results from a. through e. to those you get from using the program SelAction. Exercise 2: Pseudo BLUP EBV with the Bulmer effect a. Use BLUPEBV.XLS and compare accuracy and index weights with and without Bulmer? You can also evaluate this by changing the % selected and see how this changes index weights. b. Evaluate reduction in accuracy due to Bulmer with different heritabilities c. How does the variance of parental EBV change with the proportion selected? d. Calculate the accuracy of a parental average EBV with and without selection (Bulmer correction) for different proportions selected. Look also at the variance of parental EBV. PA = ½ EBVsire + ½ EBVdam Var(PA) = ¼Var(EBVsire) + ¼Var(EBVdam) Accuracy = sqrt(Var(PA)/VA) Exercise Day1 9 Matrix calculations using Excel You can do some basic matrix calculations with MS Excel. First put in the values of your matrices To multiply two matrices: - select an area of the size of the resulting matrix - type: =MMULT( - select the area of the first matrix - type a comma (,) - select area of the second matrix - type a close bracket ) - press: Ctrl_Shift_Enter To add or subtract a matrix (vector): - select an area of the size of the resulting matrix - type: = ( - select the area of the first matrix - type a + or - select area of the second matrix - type a close bracket ) - press: Ctrl_Shift_Enter To invert a matrix: - select an area of the size of the resulting matrix - type: =MINVERSE( - select the area of the first matrix - type a close bracket ) - press: Ctrl_Shift_Enter To transpose a matrix (vector): - select an area of the size of the resulting matrix - type: =TRANSPOSE( - select the area of the first matrix - type a close bracket ) - press: Ctrl_Shift_Enter A more specialized matrix calculation program is MATLAB. It contains many more matrix functions and mathematical function than excel. MATLAB allows you to make and run programs, draw graphs, and run simulation). A MATLAB student version is very well suitable for animal breeding problems and quite easy to use. Exercise Day1 10