Northern protocol
advertisement

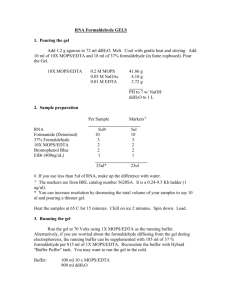
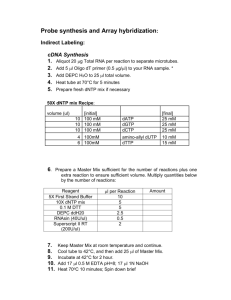
NORTHERN PROTOCOL RNA HARVESTING & SAMPLE PREPARATION (NOTE: If concerned about RNAse contamination of pipettors, the shafts can be soaked in chloroform) 1. Remove media 2. Add 500-700µL RNAzolB (mireya's fridge) per 60 mm dish (500 or less for well of a 6-well). Scrape cells off dish into Eppy. 3. Add 1/10 V chloroform 4. Mix vigorously (shake by hand), keep on ice 5' 5. Spin in cold room 15', max 6. keep aqueous phase (top) - the bottom phase is purple. Make sure not to take the white junk along, use a P200. 7. Add equal V isopropanol, mix 8. keep on ice 30' 9. spin max, 15' cold room 10. wash in 70% EtoH STORE HERE AT -20 IF YOU WILL NOT USE RIGHT AWAY 11. spin 15', cool room max speed 12. resuspend in 20µL DEPC ddH2O to spec or 20µL loading buffer to use 13. If you spec, then reppt RNA with 1/10V NaOAc pH 5.2, 2X V 100% EtOH on ice 30', spin at 4° 30', wash in 70% EtOH as above. 14. For spec, 1 OD = 40µg/mL 15. Before loading, heat samples at 65°C for 10' and cool on ice. ALTERNATIVE SAMPLE PREPARATION Sample Buffer (2X Denaturing Buffer): 50 ul formamide 18 ul 37% formaldehyde (~ 2.2 M) 10 ul 10X MOPS buffer Loading Buffer (Add 1 ul EtBr to 100 ul loading buffer) 10 mM EDTA Ph 8.0 0.25% (w/v) bromphenol blue 0.25% (w/v) xylene cyanol 50% (v/v) glycerol RNA sample preparation: < 8 ul RNA sample + 10 ul sample buffer Heat @ 65 C for 10 min (don’t forget to heat the samples) Cool on ice immediately Add 2 ul loading buffer (w/ EtBr) Load on gel FORMALDEHYDE GEL 1. Treat 250mL flask and a stir bar, gel box and combs with Rnase Zap and rinse with DEPC water 3. For 1.2% gel: 1.08g agarose, 65 mL DEPC ddH2O. Heat in microwave to dissolve 4. Add stir bar and stir in hood until 65°C (can hold in hand) 5. Add 9mL 10X MOPS, 16mL 37% formaldehyde (jug under the hood near tc room). 6. Swirl to mix and pour into gel box in the hood. Not too thick! 7. Leave in hood until cool, then can move to bench. 8. Running buffer is 1X MOPS (DEPC ddH2O, about 600mL for medium box) 9. Load samples, load dye in the first lane for tracking just use DNA SB in loading buffer (or you can leave this out if you are using the alternative sample preparation). Record order! 10. Run about 70-100V (takes 2.5 -3 hours for dye to reach the bottom at 70V) 11. Clean a plastic container very well with Rnase Zap and ddH2O. Wash gel 6X fresh ddH2O each for 5 min. 12. Wash gel 20X SSC for 30'. 13. Assemble turbo blotter (Jay's bench, paper in cabinet near Wen's bench) according to the manufacturer's directions. (wash with Rnase Zap and ddH2O then dry, then 20 #4s, 3 #2s, 2 #2s soaked in 20X SSC, the Hybond nylon membrane cut to gel size, the gel face up NO BUBBLES, then line the area around the gel with parafilm so the transfer won't short circuit, then more #2s in 20X SSC, then the wick, with about 100mL 2X SSC in the buffer chamber.) Make sure to line Hybond nylon with parafilm so you do not short circuit transfer. 14. Wrap turbo blotter in saran wrap and transfer O/N. BLOTTING 1. Next am, take down transfer. Mark position of lanes at the top with pencil. Cut upper left corner. 2. Wet only the back side of blot in 2X SSC by floating it on the top of a container of SSC 3. Place on saran wrap face up and U/V crosslink 4' (room with phosphorimager). 4. Rinse in 2X SSC 5. 5' in 1N acetic acid (reuse this solution) 6. 5' in methylene blue stain (reuse this solution). 7. destain with ddH2O from the spigot. 8. Wrap blot and xerox to document rRNA bands. Mark position with pencil in the dye lane. 9. Store at -80 until ready to probe. 10. To ready for probe, prehyb. 90' in 15 mL hybridization solution at 65°C in warm room rotator. PROBE 1. PCR or digest probe sequence 500-1000 bp, you'll need 25-50ng per probe 2. Mix 25-50ng DNA into TE for a final volume of 45µL 3. Denature 95-100°C for 5 minutes. 4. Cool on ice for 2 minutes 5. Centrifuge briefly 6. Get a "Ready to Go" probe tube from Mireya's RNAse free cabinet (Pharmacia) 7. Make sure white pellet is at the bottom of the tube. 8. Add the 45µL denatured DNA and dissolve pellet. Pipette carefully to avoid bubbles. 9. Add 5µL fresh (preferably) 32P-dCTP (NEN, Easytides, 50µCi) 10. dCTP is green, pipette to make uniform solution. 11. Place in radioactive box in warm room (37°) for 5-30 mintues (I've been doing 30). 12. Prepare column (from Bob’s shelve above the micro-centrifuge) by vortexing to resuspend the resin (not the white band). Slightly open and snap off the bottom of the column (snap HARD at score) 13. Place tube in open screw cap tube and spin 3K, 1' to clear column. Dispose of tube, move column to new tube. 14. Add the probe to the top of the column. Spin 3K, 2'. Green color should stay behind. 15. Dispose of column, put screw cap on the tube. 16. Remove 1µL of the probe and place pipette tip in small scintialltion vial. You do not need to add scintillation fluid. Count in the scint. counter using the user 10 card and program. Do "count single rack." You expect 200000-400000 counts for that 1 µL. 17. BOIL THE PROBE 5' at 95-100 °C, cool on ice 3-5'. CHURCH-GILBERT BLOTTING 1. will need to prehyb the blot in 15mL hybridization solution at 65°C in rotator over for 90min. Bottles for hybridization are on sink nearest tc room, prepare by washing several X in ddH20. These are stored in count off. 2. Add 2x1,000,000 cpm/mL hyb solution in 10mL that is 2x10,000,000. BOIL THE PROBE BEFORE ADDING TO THE HYB MIX. 3. Hybridize at 65°C O/N 4. Dispose of probe in radioactive waste 5. Rinse blot briefly with 50mL wash solution heated to 65° and dump down sink (record!) 6. Wash 3X 30' at 65° in rotator oven. 7. Wrap blot in saran wrap and expose to PI screen (2hrs - O/N) or film with screen at 80°C for O/N - several days. ( ALTERNATIVE - QUIK HYB BLOTTING) 1. Add 100µL Stratagene salmon sperm DNA to probe before boiling. Boil 4-5', place on ice 2. Prehyb in 3mL Quik Hyb solution (Strategene - in Yi's fridge) > 15' at 65°C 3. Add probe to hyb in the 3mL (still 2000000 counts/mL). Hyb 90', 65° 4. wash twice 0.1% SDS, 2X SSC 15' RT 5. wash once 0.1X SSC. ).1% SDS 55-60°, 15' (To heat solution, nuke 1') STRIPPING 1. Heat 100-150mL 1% SDS in TE to boiling in microwave 2. Add to blot and shake at RT for 30' 3. Discard wash and monitor radioactivity. 4. Repeat until no signal detected 5. Rinse with 2X SSC and prehyb again. RECIPES Loading Buffer: 50µL formamide 18µL formaldehyde 12µL H20 10µL 10X MOPS DEPC water: DEPC is in the cold box, left side near the bottom Add 1:1000 to fresh ddH2O from the spigot in 1L bottles (good to make about 2L to start) Keep at 37°C in warm room for several hours to O/N, then autoclave. 10X MOPS DEPC Dissolve 6.8g anhydrous NaOAc in 800mL H20 pH to 7.0 with 10N NaOH add 20mL 0.5M EDTA pH 8.0 DEPC treat 37°C O/N (1:1000) autoclave add 41.93g MOPS recheck pH (7.0) fill to 1L with DEPC H20 filter sterile 0.22µm filter. cover in foil, store dark. Use only until straw colored 20X SSC: in 400 mL ddH2O: 87.65 g NaCl 44.1g solium citrate (or 50.25g Na citrate 2H2O) adjust pH to 7.0 with a few citric acid crystals. adjust V to 500mL Methylene Blue 0.04% in 0.5M NaOAc pH 5.3 Acetic Acid glacial acetic acid is 17.4N, dilute with ddH20 to 1N Church-Gilbert Hybridization solution: for 50mL: 7.4mL ddH2O 25mL 1M NaHPO4 pH 7.2 17.5 mL 20% SDS 0.1mL 0.5M EDTA pH 8.0 0.5g BSA (Sigma 7906) Church-Gilbert Wash Solution: for 500mL: 450mL ddH2O 20mL 1M NaHPO4 pH 7.2 25mL 20% SDS 1mL 0.5M EDTA pH 8.0