phylotreePlugin
advertisement

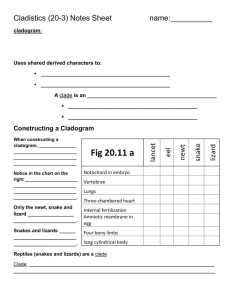
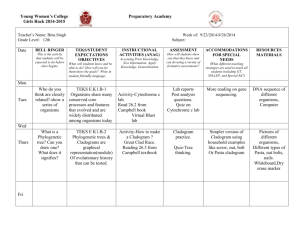
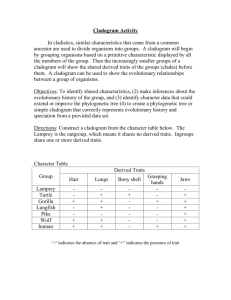
Cytoscape Phylogenetic Tree Plugin v1.0 Author: Chinmoy I.S. Bhatiya Timeline: Developed during Google Summer of Code 2009 (May - August 2009) The phylogenetic tree plugin for Cytoscape allows users to visualize phylogenetic trees in the Cytoscape environment. Trees can be imported into Cytoscape and manipulated like any other network. Information stored in the tree can be accessed using the network, node and edge attributes. Supported formats: Currently the plugin can import phylogenetic trees that are in the Phylip/Newick format and the phyloXML format. Details about these formats can be obtained from their webpages (http://evolution.genetics.washington.edu/phylip/newicktree.html) and (http://www.phyloxml.org/) respectively. Layouts: The plugin provides the following layouts for displaying phylogenetic trees: Cladogram Layouts – A cladogram is a tree laid out independent of phylogenetic distances between its elements, or in a way that all internode distances are the same. Rectangular Cladogram: A cladogram with rectangular edges. Slanted Cladogram: A cladogram with straight edges. Radial Cladogram: A cladogram in which nodes are arranged along circles of different radii. Circular Cladogram: A radial cladogram with rectangular edges. Phylogram Layouts – A phylogram is a tree laid out in a way that the distance between the nodes represents their phylogenetic distance. Rectangular Phylogram: A phylogram with rectangular edges. Slanted Phylogram: A phylogram with straight edges. Radial Phylogram: A phylogram in which nodes are arranged along circles of different radii equal to their phylogenetic distance from the root. Circular Phylogram: A radial phylogram with rectangular edges. The algorithms for drawing trees under the different layouts were adapted from a recent publication about drawing rooted networks. (Huson, 2009). The phylogram layouts position nodes in the network view based on the distances defined in the tree. It is highly recommended to use the phylogram layouts only for phylogenetic trees that have branch lengths defined, otherwise they may not be easy to visualize. If edges in the layout appear too close together or too far apart, they can be scaled by entering an appropriate value for the edge scaling factor under Settings in the layout menu. Visual Styles: The plugin provides two visual styles under the VizMapper designed specifically for trees. phylotree_DepthwiseColor colors nodes and edges in the tree in a continuous gradient reflecting the number of ancestors between the node and the root of the tree. The lighter the color of the node, the greater the number of ancestors existing between it and the root. phylotree_DepthwiseSize also colors nodes and edges similar to the phylotree_DepthwiseColor. Additionally, it also changes the sizes of nodes to reflect the number of ancestors between the node and the root of the tree. The smaller the node, the greater the number of ancestors existing between it and the root. Usage: To import a phylogenetic tree into the Cytoscape, go to "Phylogenetic tree..." under File->Import. Select the format of the tree to be imported and click on "..." to browse to the file. Please ensure that the format selected matches the format of the file selected. Click OK to begin importing the file. The tree is a valid tree, it will automatically be displayed with the slanted cladogram layout and the phylotree_DepthwiseSize layout. You may select a new layout from the Layout->Phylotree Layouts menu. In order to change visual style, click on the VizMapper tab on the panel to the left and select a visual style from the drop down list. Troubleshooting: 1. The tree imports without error but the network view displays only a black screen. - The tree is actually being displayed correctly, but because of its large size, the view has been zoomed out considerably. You may try zooming into the try or selecting a different visual style if you would prefer to visualize the entire tree at the current zoom level. 2. Nodes overlap in the phylotree_DepthwiseSize visual style. - This can occur if nodes are placed too close to each other. This problem can be remedied by changing the scale of the network view from the Layout->Scale menu. Contact: Please contact chinmoy.bhatiya@gmail.com with questions and suggestions. References: - Huson, D.H. 2009. Drawing Rooted Phylogenetic Networks. IEEE Transactions on Computational Biology and Bioinformatics. 6: 103 – 109.