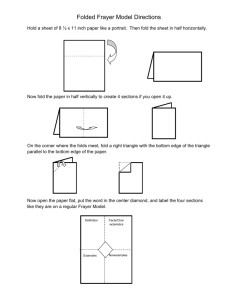
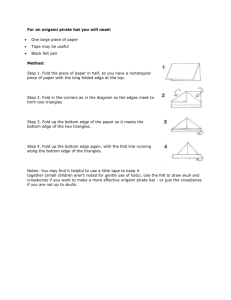
Method for phospho-peptide quantitation of MS peaks using 18O
advertisement

Robust MS Quantification Method for Phosphopeptides using 18O/16O Labeling Claus A. Andersen, Stefano Gotta, Letizia Magnoni, Roberto Raggiaschi, Andreas Kremer and Georg C. Terstappen Siena Biotech SpA, Discovery Research, Via Fiorentina 1, 53100 Siena, Italy Supporting material Full labeling Mixt labeling DpSDDEEEVVHVDR KBackground 7000 TLight KBackground T 4500 TLight T 4000 TMixt 3500 THeav y 3000 S [Observed] Background Light 6000 TMixt S [Observed] 1000 Spectral intensity Mixt THeav y 1500 Fold change = NaN [NaN NaN] 5000 AKTDHGAEIVYKpSPVVpSGDTpSPR K T 5000 Heavy S [Observed] 4000 3000 Spectral intensity 2000 Spectral intensity Direct DLpSLEEIQKK No labeling Lab.Eff = 0.85 Fold change = 1.02 [-1.1 1.2] Fold change = -Inf [NaN NaN] 1000 1000 500 0 0 S1 S2 S3 S4 S5 S6 m/z [1 Da per step] S7 0 S0 S8 Fold change = -Inf [NaN NaN] 1000 800 S2 S4 S5 S6 S3 m/z [1 Da per step] S7 S8 S0 S1 9000 DpSDDEEEVVHVDR K TLight 8000 T TMixt 7000 T THeavy S [Observed] 600 400 S2 S3 S4 S5 S6 m/z [1 Da per step] S8 3500 AKTDHGAEIVYKpSPVVpSGDTpSPR KBackground 3000 TLight Background Light TMixt Mixt T Heavy 6000 S7 Fold change = NaN [NaN NaN] Lab.Eff = 0.85 Fold change = 1.02 [-1.1 1.2] KBackground Spectral intensity DLpSLEEIQKK S1 S [Observed] 5000 4000 3000 Spectral intensity S0 Spectral intensity 2000 1500 2000 500 Inverted 2500 2500 THeav y S [Observed] 2000 1500 1000 2000 200 500 1000 0 0 S0 S1 S2 S3 S4 S5 S6 m/z [1 Da per step] S7 S8 0 S0 S1 S2 S3 S4 S5 S6 m/z [1 Da per step] S7 S8 S0 S1 S2 S3 S4 S5 S6 m/z [1 Da per step] S7 Figure S1 The degree to which a peptide is labeled (the labeling efficiency) varies dramatically between peptides. On the left is a special case where a peptide has been fully labeled (full labeling). This can be immediately seen since the peptide was absent from the treated sample, by comparing the direct and inverted experiments (label swapping). One peptide was not labeled (No labeling) and most peptides receive a partial labeling (see Figure 2b). 17-Feb-16 3:28:01 AM 1 S8 80 70 mean = 1.28 median = 1.12 stddev = 0.40 Relative occurrence 60 mean = 1.00 median = 1.00 stddev = 0.86 50 40 30 20 10 0 -6 -4 -2 0 2 4 6 Estimated fold change Figure S2 The estimated fold change is compared between the experimental spectra (red) and the simulated spectra (blue). The simulated spectra were generated as a symmetrical distribution with a standard deviation twice as large as the ones observed to allow the investigation of high fold change peptides. The experimentally observed spectra pair with a completely absent treated peptide was excluded since its theoretical fold change was –infinite. 17-Feb-16 3:28:01 AM 2 Fold change error: average(|true - estimated|) 0.9 noise [0.01 0.06] noise (0.06 0.10] 0.8 noise (0.10 0.15] noise (0.15 0.20] 0.7 noise (0.20 0.24] 0.6 noise (0.24 0.29] noise (0.29 0.34] 0.5 0.4 0.3 0.2 0.1 0 0.4 0.5 0.6 0.7 0.8 0.9 1 Estimated labeling efficiency Figure S3 When the labeling efficiency decreases the fold change error was found to increase. For a low noise level (noise [0.01 0.10]) we found an almost constant or slowly increasing fold change error upon decreasing labeling efficiency, while for increased noise level the fold change error was found to increase steadily with poorer labeling efficiency. 17-Feb-16 3:28:01 AM 3 1 noise [0.01 0.06] noise (0.06 0.10] noise (0.10 0.15] noise (0.15 0.20] Imposed labeling efficiency 0.9 noise (0.20 0.24] 0.8 noise (0.24 0.29] noise (0.29 0.34] 0.7 0.6 0.5 0.4 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 Estimated labeling efficiency Figure S4 The labeling efficiency estimate is generally quite reliable, but depends on the level of noise in the spectra. An interesting aspect is that on average it is overestimated for high labeling efficiencies and underestimated when it is low. The black line illustrates the ideal labeling efficiency. 17-Feb-16 3:28:01 AM 4 Fold change: 1.11 [1.03 1.20] Fold change error: average(|true - estimated|) KBackground S3 S4 S5 S6 m/z [1 Da per step] S8 TLight Inverted Inverted TMixt THeavy S [Observed] 0.4 S2 S3 S4 S5 S6 S7 0.45 0.3 0.1 S1 0.2 5 0.05 0.5 0.02 S8 0.45 0.4 0.6 0.2 S0 0.1 0. 2 S7 KBackground 0.4 45 0. 0.6 0.6 0.7 0.7 0.8 0.6 0.7 0.8 0.9 1 2 1.1 11.3. 1.4 0.04 0.5 0.5 0. 5 S2 0.35 0. 0.3 4 5 S1 3 0. 0.3 S0 0.7 0.150.1 0.2 5 Estimated labeling efficiency S [Observed] 0.3 0.35 0.15 0.8 0.2 TMixt THeavy pTPSLPTPPTR 5 0.2 0.9 TLight Direct 0.2 0. 0. 0.3 6 5 00.4 0 .4 .35 5 pTPSLPTPPTR 0.8 0.9 1 1.1 0.9 1 1.1 1.2 1.13.4 1.5 1.6 0.06 0.08 0.1 0.12 0.14 1.2 1.3 1.4 1.5 1.6 7 1. 0.16 0.18 Error/Signal Fold change: 2.49 [2.05 3.19] ASGQAFELILpSPR KBackground Direct Fold change: -1.01 [-1.7 2.5] TDHGAEIVYKpSPVVpSGDTpSPR KBac kgr ound Direct TLight TLight TMixt TMixt THeavy THeavy S [Observed] S [Observed] S0 S1 S2 S3 S4 S5 S6 ASGQAFELILpSPR S7 S8 KBackground S0 S1 S3 S4 S5 S6 TMixt THeavy S [Observed] S [Observed] S2 S3 S4 S5 S6 S7 S8 TLight TMixt THeavy S0 S1 S7 KBac kgr ound Inverted TLight Inverted S2 TDHGAEIVYKpSPVVpSGDTpSPR S8 S0 S1 S2 S3 S4 S5 S6 S7 S8 Figure S5 Examples of experimentally observed spectra pairs are shown in relationship to the contour plot delineating the average fold change error given the Error/Signal-ration and labeling efficiency estimated by the method presented. 17-Feb-16 3:28:01 AM 5