Determining the Size of an Unknown DNA Fragment Using Excel™
advertisement

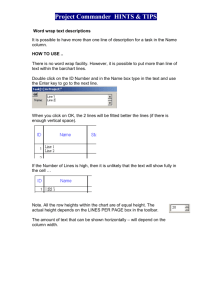
Determining the Size of an Unknown DNA Fragment Using Excel™ 1. Label columns A-C as “base pairs” (or “bp”), “Distance (in cm)”, and “log10 bp”. 2. Enter the size of the standard fragments in column “A”. Enter the distance traveled by the corresponding markers fragments in column “B”. 3. Click on the first box in the “log bp” column. Click “InsertFunction” or click on the “fx” icon on the toolbar. In the category selection box, select “Math and Trig”. Then select the “LOG10” function from the list. The “LOG10” dialog box will appear. Drag the “LOG10” dialog box to a convenient location, and then click on the first entry in the “bp” column. The log10 of the value in column “A” will appear in column “C”. Click on the lower right corner of the box in column “C” containing the new value and drag it down the entire column. The log value corresponding to the base pair values in column “A” will appear in column “C”. 4. Select the values (not the headers) in columns “B” and “C”. Click on the Chart Wizard icon on the toolbar. Select “XY scatterFinish”. A chart will appear over the spreadsheet representing the data in the highlighted range. 5. Now A “Chart” button should appear in the menu bar at the top of the page. Click “ChartAdd trendline”. In the popup select “Linear Trend/Regression”. Click on the “Options” tab in the window and check the boxes labeled “Display equation on the chart” and Display r-squared value on the chart”. Click “OK”. The best fit should be on the chart. Notice that the linear regression equation (y=mx+b) and the r-squared value are superimposed on the graph near the line. The closer the r-squared value is to “1”, the higher the probability that all of the points lie on, or very close to, the tyrendline. For clarity, you may wish to label the chart axes using the “Chart options”. 6. To solve for the size of your unknown fragment, start a new column labeled “distanceunknowns” or a label of your choosing. Enter the distance traveled by the fragments in centimeters. 7. Label a new column “m * distance”. Click “Insertfunction”, and again select the “Math and Trig” group. Select “Product” in the function name category. 8. In the product dialogue box, enter the regression equation “m” value (from y=mx+b) in the top box. In the second box, enter the value of the first distance traveled. 9. Select a box to the right of your first entry (label the column “SUM(mx+b)”)and again insert a “Math and Trig” function, this time the SUM function. Click OK. In the dialogue box, enter the mx product calculated in step 8 in the top box, and enter the “b” value from the regression equation in the second box. Select “OK”, and the sum will appear in the highlighted box. Drag the function down the column until all sums are calculated. 10. At this point, you now have a value for y. Remember that the distance between bands on an agarose gel changes logarithmically. Thus, the value you now have is the log10 of the base pairs. To get the tru number of base pairs you need to take the antilog of this value. Thus, 11. Labe a new column (Ex., “unknown bp values”). Highlight the box beneath the label and click “InsertFunctionMath and TriPOWER”. Enter 10 in the box labeled “Number” in the dialogue box (because this is log base 10). In the “POWER” box, enter the value from the corresponding cell in the “SUM(mx+b)” column. Click OK. The number of base pairs of you fragment should have been calculated. Check to see if it makes sense. If so, drag the first cell down the rest of the column and record your values. 12. Compare these values to your estimates derived from using the semi-log paper.