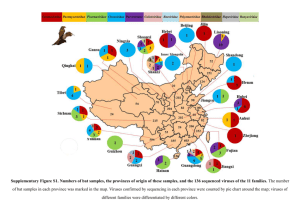
Supplementary Figures Titles Supplementary Figure 1. Vertebrate
advertisement

Supplementary Figures Titles Supplementary Figure 1. Vertebrate CYP2 phylogenetic analysis. Detailed phylogenetic analysis of the CYP2 proteins in vertebrates; a total of 24 CYP2 subfamilies were included. A JTT+I+G (Jones Taylor Thornton + invariant sites + gamma distribution) model was selected using ProtTest (Abascal et al. 2005). A maximum likelihood approach (RAxML) (Stamatakis et al. 2008) was applied to the final alignment of 196 sequences to determine the phylogenetic history. Bayesian inference was computed based on the final CYP2 alignment. All nodes are labeled with bootstrap values and posterior probabilities. Accession numbers are provided for opossum sequences, as these lack full gene nomenclature. Accession numbers for all other sequences are provided in Supplementary Table 2. Supplementary Figure 2. Phylogenetic analysis of the CYP2J / 2P/ 2N / 2AD / 2AE / 2V / 2Z cluster. A phylogenetic analysis of the CYP2J-2Z cluster (node 7 in Figure 1) based on a maximum likelihood approach is shown. Bootstrap values and posterior probabilities are labeled on all nodes. The CYP2 alignment included 343 amino acids for this analysis. Ciona CYP2N, CYP2J and CYP2X sequences were used as outgroups. Accession numbers are provided for opossum sequences, as these lack full gene nomenclature. Accession numbers for all other sequences are provided in Supplementary Table 2. Supplementary Figure 3. Phylogenetic analysis of the CYP2C clade. A phylogenetic analysis of the CYP2C cluster (node 2 in Figure 1) based on a maximum likelihood approach is shown. Bootstrap values and posterior probabilities are labeled on all nodes. The CYP2 alignment included 346 amino acids for this analysis. The outgroup sequences were from the CYP2E (mammalian) subfamily. Accession numbers are provided for opossum sequences, as these lack full gene nomenclature. Accession numbers for all other sequences are provided in Supplementary Table 2. Supplementary Figure 4. Speciation patterns and the gene duplications in the CYP 2W, 2X, 2AA and 2K subfamilies. The evolution of vertebrate subfamilies are symbolized by duplication ( ) and gene loss ( ) events. Diversification of vertebrate species is symbolized by speciation patterns ( ). The two CYP2 putative sequences (Opossum CYP2 putative (XP_001369607) and Frog CYP2 (NP_001037917) are strongly supported through phylogenetic analysis and are closely clustered with CYP2W, a mammalian subfamily. Supplementary Figure 5. Speciation patterns and the gene duplications in the CYP 2A, 2Y, 2F, 2B, 2S and 2G subfamilies. The evolution of vertebrate subfamilies are symbolized by duplication ( ) and gene loss ( ) events. Diversification of vertebrate species is symbolized by speciation patterns ( ). Dashed lines represent areas of topology with weak support. Frog 2C8 is not a true 2C sequence, it does not cluster with the 2C sequences. Frog 2C8 sequence is clustered between CYP2Y and CYP2M, both are actinopterygian subfamilies. Supplementary Figure 6. Speciation patterns and the gene duplications in the CYP 2J, 2N, 2AD, 2P, 2Z, 2V and 2AE subfamilies. The evolution of vertebrate subfamilies are symbolized by duplication ( ) and gene loss ( ) events. Diversification of vertebrate species is symbolized by speciation patterns ( ). Dashed lines represent areas of topology with weak support. This subfamily cluster presented with complexity in its topology as seen with poor support for the CYP2V and CYP2AE subfamilies which are zebrafish specific. The speciation and duplication patterns were derived from the original CYP2 phylogenetic analysis (Supplementary Figure 1) and not from distinct analysis (Supplementary Figure 2).