Supporting Information - Springer Static Content Server
advertisement

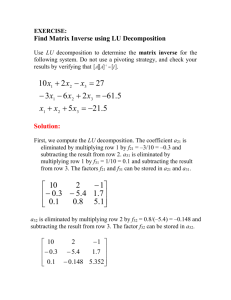
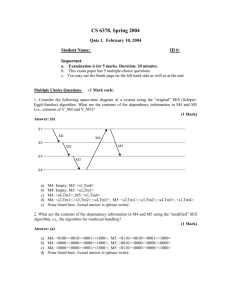
Molecular modeling of cytochrome b5 with a single cytochrome c-like thioether linkage Ying-Wu Lina, b*, Yi-Mou Wuc, Li-Fu Liaoa, Chang-Ming Niea a School of Chemistry and Chemical Engineering, University of South China, Hengyang 421001, China b State Key Laboratory of Coordination Chemistry, Nanjing University, Nanjing 210093, China c Institute of Pathogenic Biology, University of South China, Hengyang 421001, China Supporting Information Topology file of residue HOBC defined for oxidized C-type heme with one thioether linkage and bis-His coordination: PRES HOBC -0.50 !-0.73 ! Patch for HEME to C-type Cytochrome b5 links. ! Patch residues must be 1:HSD, 2:HSD, 3:CYS (C-terminal), and ! 4:HEMO. ! Modified by Ying-Wu Lin 10/3/2011 to include charge ! redistribution and to be a single patch. GROUP ATOM 1CB ! CT2 -0.09 !-0.22 ! ATOM 1ND1 NR1 -0.36 !-0.61 ! 1HD1 1HE1 | ATOM 1HD1 H 0.32 ! 0.45 ! 1HB1 ATOM 1CG -0.05 ! 0.32 ! | CPH1 GROUP ! 0.25 ! 0.24 ! ATOM 1HE1 HR1 0.13 ! 0.27 ! ATOM 1NE2 NR2 -0.70 ! -0.51 ! ATOM 1CD2 CPH1 0.22 !-0.04 ! ATOM 1HD2 HR3 0.10 ! 0.16 ! DELETE ANGLE 4NA 4FE 4NC / || ! -1CB-1CG ATOM 1CE1 CPH2 / 1ND1-1CE1 | || \\ 1HB2 || 1CD2-1NE2 | 1HD2 \ 4FE- 4NB 4FE 4ND BOND 1NE2 4FE ANGLE 1CD2 1NE2 4FE 1CE1 1NE2 4FE ANGLE 1NE2 4FE 1NE2 4FE 4NC 1NE2 4FE 4NA 1NE2 4FE 4NB 4ND IMPR 1NE2 1CD2 1CE1 4FE !add impr for heme/his planarity IC 1CD2 1NE2 4FE 4NA 0.0000 0.0000 0.0000 0.0000 0.0000 IC 1CD2 1NE2 4FE 4NB 0.0000 0.0000 0.0000 0.0000 0.0000 S1 IC 1CD2 1NE2 4FE 4NB 0.0000 0.0000 0.0000 0.0000 0.0000 IC 1CD2 1NE2 4FE 4NB 0.0000 0.0000 0.0000 0.0000 0.0000 IC 1CE1 1NE2 4FE 4NA 0.0000 0.0000 0.0000 0.0000 0.0000 ! Heme to Histidine portion of patch GROUP ATOM 2CB ! CT2 -0.09 !-0.22 ! ATOM 2ND1 NR1 -0.36 !-0.61 ! 2HD1 | ATOM 2HD1 H 0.32 ! 0.45 ! ATOM 2CG -0.05 ! 0.32 ! CPH1 GROUP ! 2HB1 | / 2ND1-2CE1 / || ! -2CB-2CG ATOM 2CE1 CPH2 0.25 ! 0.24 ! | ATOM 2HE1 HR1 0.13 ! 0.27 ! 2HB2 ATOM 2NE2 NR2 -0.70 !-0.51 ! ATOM 2CD2 CPH1 0.22 !-0.04 ! ATOM 2HD2 HR3 0.10 ! 0.16 ! DELETE ANGLE 4NA 4FE 4NC 2HE1 || \\ || 2CD2-2NE2 | \ 2HD2 4FE- 4NB 4FE 4ND BOND 2NE2 4FE ANGLE 2CD2 2NE2 4FE 2CE1 2NE2 4FE ANGLE 2NE2 4FE 2NE2 4FE 4NC 2NE2 4FE 4NA 2NE2 4FE 4NB 4ND IMPR 2NE2 2CD2 2CE1 4FE !add impr for heme/his planarity IC 2CD2 2NE2 4FE 4NA 0.0000 0.0000 0.0000 0.0000 0.0000 IC 2CD2 2NE2 4FE 4NB 0.0000 0.0000 0.0000 0.0000 0.0000 IC 2CD2 2NE2 4FE 4NB 0.0000 0.0000 0.0000 0.0000 0.0000 IC 2CD2 2NE2 4FE 4NB 0.0000 0.0000 0.0000 0.0000 0.0000 IC 2CE1 2NE2 4FE 4NA 0.0000 0.0000 0.0000 0.0000 0.0000 !(C-terminal) Heme to Cysteine (Thioether bond) portion of patch GROUP ! ATOM 3CB CT2 ATOM 3SG S 0.07 ! -3CB-3SG-4CAC-0.23 ! DELETE ATOM 3HG1 BOND 3SG 4CAC ANGLE 3CB 3SG 4CAC ANGLE 3SG 4CAC 4C3C ANGLE 3SG 4CAC 4CBC ANGLE 3SG 4CAC 4HAC DIHEDRAL 3CB 3SG 4CAC 4C3C DIHEDRAL 3CB 3SG 4CAC 4CBC DIHEDRAL 3CB 3SG 4CAC 4HAC DIHEDRAL 3CA 3CB 3SG 4CAC DIHEDRAL 3HB1 3CB 3SG 4CAC DIHEDRAL 3HB2 3CB 3SG 4CAC S2 -1 (deg.cm .dmol ) 9 Wild Type cyt b5 6 2 N57C cyt b5 3 [Theta] x 10 4 0 -3 -6 -9 200 210 220 230 Wavelength (nm) 240 250 Fig. S1 Circular dichroism (CD) spectra of WT cyt b5 and N57C cyt b5 in the oxidized state collected at 25 C from 190 to 250 nm (0.1 cm path length) with a Jasco model J720 spectropolarimeter. The protein concentration was 25 μM, dissolved in 100 mM sodium phosphate buffer at pH 7.0. (The experiment was performed by Y.-W. Lin in Prof. Zhong-Xian Huang’s group at Fudan University, China). S3 a b N57C cyt b5 X-ray:1cyo c d NMR:1hko DS cyt b5 Fig. S2 Ramachandran plots of amino acids for modeling structure of bovine N57C cyt b5 (a), X-ray structure of cyt b5 (PDB entry 1cyo) (b), NMR structure of cyt b5 (PDB entry 1hko, model 1) (c), and modeling structure of DS cyt b5 (d), as separately by dashed line. The horizontal and vertical axes are phi and psi, respectively. Protein residues are mapped to the Ramachandran diagram with yellow squares. The most allowed regions of Ramachandran space are colored blue; partially allowed regions are colored green. S4