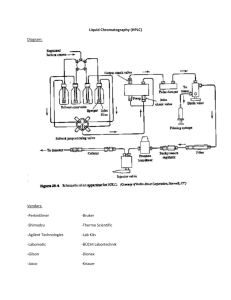
High performance liquid chromatography (HPLC) is
advertisement

Quantitative Antibody Profiling with Detailed Structural Analyses of N-Glycans: Sequential Disassembly by ITMSn Andrew Hanneman, David Ashline, Hailong Zhang, and Vernon Reinhold The University of New Hampshire Glycomics Center and Glycan Connections LLC Quantitative HPLC profiling of fluorophore tagged N-glycans is often coupled with online mass spectrometry (MS, MS2) to determine composition and partial sequence information and additional details uncovered by exoglycosidase treatments. These approaches can leave unresolved structural issues requiring further investigation particularly with samples involving glycoengineered products or nontraditional expression systems. For improved understanding about glycan structural biology including branching, linkages, and epitopes HPLC peaks may be separately collected, infused, and studied by nanospray ion trap sequential mass spectrometry (ITMSn), and including the incorporation of an additional methylation step. HPLC fractions may also be probed enzymatically prior to methylation and MSn. Direct infusion provides an enhanced duty cycle to dig deep into structural details transparent to MS2 and enzymology alone. Facile rupture of the N-linked chitobiose bond in MS2 removes the fluorescent tag and core fucosylation (if present), leaving abundant product ions to detail the remaining structure at higher CID stages (MS3-7). Fragments are confirmed by spectral matching in a library compiled from synthetic standards and well characterized natural products. Glycan structural reassembly follows from tracking fragmentation pathways, matching scars and mass values, and use of spectral documentation as a substitute for unspecified (bracketed) compositions and biological inference. The described protocols introduce an opportunity to resolve glycan structural details de novo from bioengineered products where fluorescence profiles or biological data indicate undefined or aberrant components.