UGT activity by HPLC method development and preliminary
advertisement

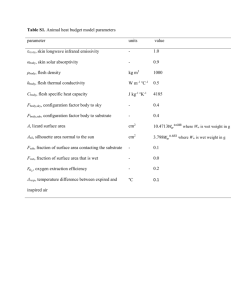
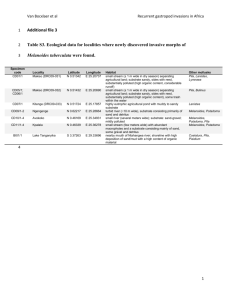
Michael Court Created June 2001; Updated November 2009 UGT activity assay by HPLC – method development and preliminary phenotyping 1. 2. 3. 4. 5. 6. 7. Develop HPLC method to measure glucuronide levels a. Check for previous method references b. Dissolve substrate and glucuronide in methanol (10 mg / 100 mL) c. Dry down substrate and glucuronide and reconstitute in 50 mM PO4 buffer d. Run UV (or fluorescent) scan of substrate and glucuronide and determine the lambda max for the analyte – set detector to this value; tune mass detector if using LC-MS. e. Set up HPLC to equilibrate i. 20 mM KH2PO4 pH = 4.5 (uncorrected pH) ii. start at 0% acetonitrile concentration iii. flow rate 1 mL/minute (pressure < 200 bar) f. Inject ~10 uL of sample g. Run acetonitrile gradient from 0% to 50% over 30 minutes (return to 0% at end) h. Optimize method (starting and end acetonitrile concentration) such that you get separation of glucuronide and other peaks and substrate in reasonable time (<40 minutes; ideally 20 minutes) i. Identify a suitable internal standard – one that separates well from substrate and product and ideally something chemically similar to substrate or product j. Check this method using a liver microsome incubation (incubate substrate with UDPGA and microsomes for up to 360 minutes) k. If no glucuronide standard available – identify potential glucuronide peak: i. Include negative controls --*No UDPGA (best) *No incubation time *No enzyme ii. Treat samples with beta-glucuronidase (see supporting protocol) – glucuronide should disappear iii. Other evidence - UV spectrum from diode array; if mass spec available - base ion mass (substrate +176) and fragmentation pattern (neutral 176 loss). Establish initial rate conditions of time, protein and alamethicin concentration. a. Do time linearity study at fixed liver microsome protein concentration (0.5 mg/mL protein – establish highest time that can be used with linear product formation. Eg. 2.5, 5, 10, 15, 20, 30, 45, 60 minutes b. Protein linearity study using the time from previous study. Eg. 0.1, 0.2, 0.3, 0.4, 0.6, 0.8, 1.0 mg/mL c. Alamethicin activation study using time and protein from previous study. i. Alamethicin stock solution = 2.5 mg / mL (in methanol) ii. Final concentration = 0.025 mg / mL (add 2.5 uL per 250 uL incubation volume) for 0.8 mg / mL protein concentration iii. 0.03125 mg alamethicin per mg protein iv. Try 0.01, 0.02, 0.03, 0.05, 0.1, 0.2 mg alamethicin per mg protein Perform preliminary Km finding study using pooled HLMs a. Eg. 1, 2, 5, 10, 20, 50, 100 uM (if Km = 10 uM) b. Need to have 2 – 3 points below final Km and up to 10x Km c. Generally use 8-10 points in duplicate If have liver bank - determine activities for all HLMs (for correlation analyses and to identify fast and slow metabolizers) at ~Km substrate concentration. Determine kinetics for selected individual HLMs (eg. 3 fast, 3 slow) to determine variability between individuals and compare to recombinant enzyme values. Screen recombinant UGTs for activity (in duplicate at 100 uL incubation volume) a. Substrate concentration - HLM Km and 10x Km b. UDPGA = 5 mM; MgCl2 = 5mM. c. Protein - 0.5 mg / mL (less if nonspecific protein effect found) d. Time - up to 6 hours for screen - reduce for final data. e. Alamethicin - same as for HLMs Determine kinetics for most active isoforms and compare with HLMs. Michael Court Created June 2001; Updated November 2009 Generic protocol for UGT activity assay 1. Add 100 uL of UGT substrate (and inhibitor if assessing inhibition) dissolved in methanol to empty “incubation tubes” (0.5 or 1.5 mL polypropylene microcentrifuge) and dry down in a vacuum oven (set at 25-45°C depending on heat stability; can also do in speed vac). 2. Prepare “UDPGA cofactor solution” on ice in a microcentrifuge tube. For each 100 uL incubation volume add: a) 0.645 mg UDPGA powder (5 mM final). b) 10 uL 50 mM magnesium chloride in water solution (5 mM final). c) 10 uL 50 mM saccharolactone in water solution (optional). d) 25 uL 100 mM potassium phosphate in water buffer, pH 7.5. e) Balance to 50 uL with water and vortex. 3. Place incubation tubes on ice and add 50 ug of UGT enzyme source (HLM protein or recombinant protein), 2.5 ug alamethicin (2.5 ug / uL methanol; 50 ug alamethicin / mg microsomal protein) and balance to a volume of 50 uL with 50 mM potassium phosphate buffer, pH 7.5 (0.5 mg protein / mL final concentration). 4. Preincubate tubes at 37° for 5 minutes. 5. Start reaction by adding 50 uL of UDPGA cofactor solution, mix by gently flicking the tube (do not vortex rUGTs), cap tube, and incubate for up to 6 hours. 6. To aid in identification of the glucuronide metabolite peak also include 3 negative controls that: a) contain no UDPGA; b) contain no substrate; and c) are not incubated, immediately treated with stop solution, vortexed and centrifuged. 7. Stop reactions with 100 uL of ice cold acetonitrile with 5% acetic acid, vortex, and centrifuge at 14,000g for 10 minutes. 8. Transfer 190 uL to glass HPLC vials, dry down in a vacuum oven and reconstitute with 95 uL of water. 9. Analyze 10 - 50 uL of the incubate by HPLC.