TPJ_3002_sm_AppendicesS1S2
advertisement

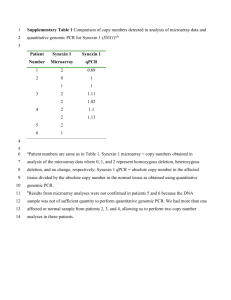
Supplementary material for Bonaventure et. al. Supplementary results: Analysis of TPC1 mRNA expression To investigate the possibility that the onset of the fou2 phenotypes correlate with changes in the level of TPC1 mRNA, the expression of this transcript was analyzed in mutant and wt leaves at different stages of development by 2 different methods. Gel blot analysis did not detect changes in the steady-state levels of TPC1 transcripts in young and mature wt and mutant leaves (Fig. S2) and also to exogenous JA or mechanical wounding (Fig. S2). In contrast, RT-qPCR detected 2-fold higher levels of TPC1 mRNA in young wt and fou2 leaves compared to mature leaves, and 2-fold higher levels in wounded fou2 leaves but not in those of wt (Fig. S2). Thus, based on the more sensitive qPCR technique, TPC1 transcripts appear to be slightly more abundant in young leaves and wound-induced in fou2 but not in wt leaves. Supplementary Materials and Methods: Genetic mapping: The fou2 mutant was backcrossed three times with Col-0 wt to remove second site mutations. To generate a mapping population, fou2 homozygous plants (Col-0) were outcrossed to wt Landsberg erecta plants. An initial F2 mapping population of 50 fou2 homozygous individuals was genotyped with SSLP markers. After positioning the fou2 gene close to marker ciw5 on top of chromosome IV, 519 fou2 homozygous individuals from the same F2 population were genotyped using existing CAPS markers located in the same chromosomal region (Fig. 4A). After positioning the fou2 allele close to marker T5L23.3, new CAPS and SSLP markers were designed based on Col-0 and Ler polymorphisms. These new markers were named according to their BAC location (Fig. 4A, Table S2). The mutation was finally bracketed in a chromosomal segment of approximately 30 Kb, which contained seven genes and was included in BAC clones F9H3 and T5L23. Sequencing : The complete coding sequence of the genes contained between markers F9H3.3 and T5L23.3 (Fig. 4A) were PCR-amplified from fou2 genomic DNA using the LA Taq enzyme (Takara, Shiga). Direct sequencing of these genomic PCR fragments was performed. Retrieved sequences were aligned against the publicly available Arabidopsis genomic sequence. The primers used to amplify the TPC1 gene were acttggatgctcctgaacttcttgc and accaattatagccaattgagcggat. The sequences of the primers used to amplify the remaining genes in the interval analyzed are available upon request. The TPC1 cDNAs from wild-type (Col-0) and fou2 plants were obtained by reverse transcription of total RNA as described above and PCR using primers gctttcaattcgatccagtaa and gctcttaacgatcttgttttg. The mutation found in fou2 plants was confirmed by sequencing the TPC1 gene and its corresponding cDNA from fou2 and wild-type plants (Col-0). No mutations were identified in the sequences corresponding to the remaining genes analyzed. Sequence comparison of various TPC1 putative orthologs was conducted with ClustalW software. Vector construction and plant transformation: A segment of genomic DNA spanning the complete TPC1 coding sequence and its own promoter was PCR amplified using LA Taq enzyme (Takara, Shiga) and primers (Table S3) containing gene specific sequences and Gateway cloning sites attB1 and attB2 (Invitrogen, CA). The 7 kb genomic amplicon was cloned into the binary vector pMDC100. The TPC1 cDNA was PCR amplified using LA Taq enzyme (Takara, Shiga) and primers (Table S3) containing gene specific sequences and Gateway cloning sites attB1 and attB2 (Invitrogen, CA). The 2.2 kb genomic amplicon was cloned into the binary vector pMDC32. Arabidopsis fou2 and wild-type (Col-0) plants were transformed by Agrobacterium-vacuum infiltration. Transgenic seedlings were selected on agar plates containing kanamycin (50 g/ml) or hygromycin (25 g/ml). Northern blot analyses: Total RNA extraction and Northern blots were performed using standard procedures. A 550 bp PCR fragment derived from amplification of the At-TPC1 cDNA was radiolabeled with [-P32]-dCTP by primer extension and used as a probe for filter hybridizations. T-DNA insertion lines: Homozygous plants containing a T-DNA insertion in TPC1 (SALK_145413 and GABI865C03) were identified by PCR using gene and T-DNA specific primers (Table S4). Both T-DNA/TPC1 junctions of SALK_145413 line were amplified and sequenced to confirm the T-DNA insertion point. Total RNA was extracted from leaves of both homozygous ko lines, reversed transcribed and the presence of full length ATPC1 mRNA evaluated by PCR (primers: gctttcaattcgatccagtaa, gctcttaacgatcttgttttg). The eIF4A1 mRNA was co-amplified as a control using the same primers as indicated above. Mutant crosses: Pollen from fou2 plants was used to pollinate coi1-1 (Col-0) flowers. fou2 coi1-1 double mutants were retrieved in the F2 generation. The presence of homozygous coi11 and fou2 alleles was confirmed by CAPS markers. The fou2 mutation destroys an EcoRV site present in wt genomic DNA and a CAPS marker was developed (primers: cttatcacaacaactgagagcc, cttcagagccatctccaacac). A CAPS marker for the coi1-1 allele has been previously described (15). Heterozygous seedlings derived from the F1 generation of reciprocal crosses between fou2 and tpc1-2 were analyzed with CAPS markers (fou2) and PCR (T-DNA) (see above).